·Letter to the Editor··Current Issue· ·Achieve· ·Search
Articles· ·Online
Submission· ·About IJO·
Homozygosity
mapping of a
consanguineous Pakistani family affected with oculocutaneous albinism to Tyrosinase gene
Muhammad Shakil1, Muhammad Ikram Ullah2,
Shabbir Hussain1, Sabika Firasat3, Saqib Mahmood4,
Haiba Kaul1
1Department of
Biochemistry, University of Health Sciences, Lahore 54600, Pakistan
2Department
of Biochemistry, Faculty of Biological sciences, Quaid i Azam University,
Islamabad 45320, Pakistan
3Department
of Animal Sciences, Quaid i Azam University, Islamabad 45320, Pakistan
4Department of Human
Genetics and Molecular Biology, University of Health Sciences, Lahore
54600, Pakistan
Correspondence
to: Haiba
Kaul. Department of Biochemistry, University of Health Sciences,
Lahore 54600, Pakistan. haibakaul@gmail.com
Received:
2014-12-17
Accepted: 2015-09-07
DOI:10.18240/ijo.2016.05.28
Citation:
Shakil M,
Ullah MI, Hussain S, Firasat S, Mahmood S, Kaul H.
Homozygosity
mapping of a consanguineous Pakistani family affected with oculocutaneous
albinism to Tyrosinase gene. Int
J Ophthalmol 2016;9(5): 794-796
Dear Sir,
I am Haiba
Kaul, from
the Department of Biochemistry, University of Health Sciences,
Lahore,
Pakistan. I write to present a study of oculocutaneous albinism (OCA) in
consanguineous Pakistani families.
OCA is a genetic defect of melanin biosynthesis that mainly
affects eyes, skin and hair. It is a congenital condition and the affected
individuals have reduced or completely absent melanin pigment in their eyes,
skin and hair. Clinical manifestations of the disease included visual problems that are atypical expansion of retina and
unusual prototypes of nerve relations established in eye and brain that might
lead to visualization issues[1]. Other features include heritable nystagmus, decreased
pigmentation of iris (iris luminousness), diminished pigmentation of the
retinal epithelium, foveal hypoplasia, and compressed visual acuity[2].
In OCA,
diverse genetic heterogeneity has been documented and seven loci have been
associated with the disease (OCA1-7). Among these loci, four
genes: tyrosinase (TYR),
pink eyed dilution for P-protein (P), tyrosinase-related protein (TYRP1),
and solute carrier 45 subunit A2 (SLC45A2) are well documented to cause different
types of OCA type (OCA1-4) respectively.
Mutations in two novel genes SLC24A5[3] and C10orf11[4] are recently discovered resulting
into OCA-6 and OCA-7 respectively. OCA-5 locus has been discovered but its gene
is yet to be identified[5]. Of the various types of OCA, OCA1 (MIM 203100) results
due to mutations
in TYR (MIM 606933) which resides on chromosome 11q14.3[6]. Mutations that resulted in complete
lack of TYR activity are known as
OCA1A, whereas mutations retaining some enzyme activity outcome in another type
of albinism called OCA1B.
Due to the strong consanguinity culture in Pakistan, diseases
segregating in recessive mode are quite common and thus the incidence of
albinism is much greater in our population as compared to the
non-consanguineous populations. Till today, very few studies
are being conducted to explore OCA
genes harboring in Pakistani families. In a family study, mutations were
reported in TYR alleles and in TYRP1 genes in Pakistani patients[7]. Recently,
a study reported novel locus, OCA5, in a consanguineous Pakistani family[6].
This study was undertaken with the aim to decipher the genetic basis of
OCA in consanguineous Pakistani families using linkage analysis approach. Prior to the start of this
study, ethical approval was taken from institutional review board
(IRB) of the
University of Health Sciences, Lahore, Pakistan. We enrolled ten families
affected with OCA which belonged to the Punjabi ethnic group
with at least
two affected in each kindred. Affected members of the enrolled families were
physically and clinically examined at the Layton Rehmatulla
Benevolent
Hospital, Lahore, Pakistan. Blood samples in EDTA containing vacutainers
were
collected from the affected and unaffected members of the enrolled families. Genomic DNA was extracted from
all the samples collected using
a modified phenol chloroform method as described by Kaul et al[8]. Genotyping was carried out by using
microsatellite markers for four known OCA loci (OCA1-OCA4). These include (TYR, OCA2, TYRP1 and SLC45A2). Highly polymorphic short tandem
repeat (STR) markers were selected from Marshfield maps
(http://www.marshfieldclinic.org/research/pages/index.aspx)
and the National Center for Biotechnology Information
(http://www.ncbi.nlm.nih.gov/). These primers were commercially synthesized with
forward primers labeled with fluorescent FAM dye. Polymerase chain
reaction (PCR) and linkage protocols were used according to
previous standards[9]. Analysis of specific genotypes were
assigned using
Peak Scanner™
Software v1.0 software (Applied Biosystems).
Peak scanner sizes different nucleic acid fragments that identify peaks
and fragment sizes for application specific capillary electrophoresis assays.
This data was used to construct haplotypes of the families
using Cyrillic® software. Statistical scoring using,
logarithm of odds (LOD) score was calculated to evaluate the linkage of
respective OCA locus. Two-point linkage
analysis was performed using the FASTLINK version of MLINK from the LINKAGE
Program Package[10]. An
autosomal recessive mode of inheritance with complete penetrance and a disease
allele frequency of 0.001 were used for the analysis.
Out of ten families
selected for this study, one family AL03 was mapped to TYR gene on chromosome 11q14.3. AL03
belongs to a remote village of Punjab province of Pakistan. The
family belongs to the Mughal caste and seldom marries out of the family
and thus is highly consanguineous. The pedigree was drawn up to
six generations with five affected (one deceased) individuals segregating
disease in an autosomal recessive manner (Figure 1). The affected
individuals (V-4, VI-2) were examined by medical physicians at the local
hospital. These individuals showed
phenotypes of white hair, nystagmus and
decreased visual acuity but are able to perform daily work and study with the
use of visual aid devices. Physical examination and clinical
investigations established OCA phenotype.
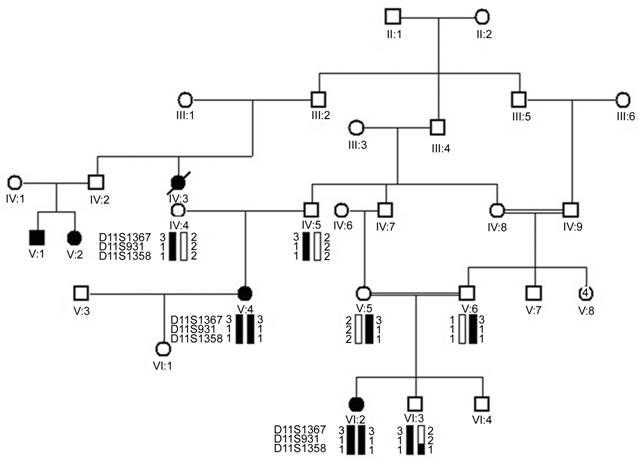
Figure 1
Haplotype of AL03 for markers (D11S1367, D11S931
and D11S1358)
spanning TYR gene Squares: Males; Circles: Females; Filled squares
and circles: Affected individuals; Blank
squares and circles: Unaffected individuals; Single lines: Non-cousin marriage;
Double lines:
Consanguinity.
Haplotype analysis
revealed that in family AL03 both the affected individuals V-4 and VI-2 were
homozygous for the alleles of three markers D11S1367, D11S931 and D11S1358 (Figure 1). The gene TYR resides between markers D11S1367, D11S931 and
D11S1358. Two-point linkage analysis was performed using the FASTLINK
version of MLINK. The highest LOD score of 1.80 (θ = 0.00) was
obtained with D11S1367 for AL03.
The results of linkage evidently showed that there is association of TYR
gene in this family.
TYR
gene
encodes TYR, copper containing
enzyme, which catalyzes conversion of tyrosine to melanin and thus is important
for melanin biogenesis[7]. Mutations
of TYR gene
have been
documented in various populations worldwide[11]. However,
molecular analysis of OCA in Pakistani families has not been carried out on
large scale. There are few reports regarding mapping of OCA genes in Pakistani
families[12]. Apart from the linkage of one OCA family, nine
other families were failed to link to any of four genes:
TYR, pink eyed P, TYRP1, and SLC45A2. This depicts that a high genetic heterogeneity is present in our
population for OCA. We can therefore conclude that the remaining families might
harbor genetic defect underlying in other 3 gene/loci. Conversely, there is a
high probability that a new gene that still remains to be identified in OCA
pathology might be responsible for the disease.
In
conclusion, this study reports a family designated as AL03 with two members
affected with OCA linked to TYR gene. None of other nine
families screened were found linked to the genes screened for OCA.
ACKNOWLEDGEMENTS
The authors are thankful to the families for their
participation in the study and the Layton Rehmatulla Benevolent Trust for help in clinical investigations. The
authors are also thankful to Higher Education Commission (HEC) Islamabad,
Pakistan for partly supporting this study.
Conflicts
of Interest:
Shakil M, None;
Ullah MI, None;
Hussain S, None;
Firasat S, None;
Mahmood S, None;
Kaul H,
None.
REFERENCES
1 Yahalom C,
Tzur V, Blumenfeld A, Greifner G, Eli D, Rosenmann A, Glanzer S, Anteby I.
Refractive profile in oculocutaneous albinism and its correlation with final
visual outcome. Br J Ophthalmol 2012;96(4):537-539. [CrossRef] [PubMed]
2 King RA,
Olds DP, Townsend D. Mechanisms of hypopigmentation in human oculocutaneous
albinism. Prog Clin Biol Res 1988;256:183-191. [PubMed]
3 Wei AH, Zang
DJ, Zhang Z, Liu XZ, He X, Yang L, Wang Y, Zhou ZY, Zhang MR, Dai LL, Yang XM,
Li W. Exome sequencing identifies SLC24A5 as a candidate gene for
nonsyndromicoculocutaneous albinism. J
Invest Dermatol 2013;133(7):1834-1840. [CrossRef] [PubMed]
4 Grønskov K,
Dooley CM, Østergaard E, Kelsh RN, Hansen L, Levesque MP, Vilhelmsen K,
Møllgård K, Stemple DL, Rosenberg T. Mutations in c10orf11, a
melanocyte-differentiation gene, cause autosomal-recessive albinism. Am J Hum Genet 2013;92(3):415-421. [CrossRef] [PubMed] [PMC free article]
5 Kausar T,
Bhatti MA, Ali M, Shaikh RS, Ahmed ZM. OCA5, a novel locus for
non-syndromicoculocutaneous albinism, maps to chromosome 4q24. Clin Genet 2013;84(1):91-93. [CrossRef] [PubMed]
6 Tomita Y,
Takeda A, Okinaga S, Tagami H, Shibahara S. Human oculocutaneous albinism
caused by single base insertion in the tyrosinase gene. Biochem Biophys Res Commun 1989;164(3):990-996. [CrossRef]
7 Forshew T,
Khaliq S, Tee L, Smith U, Johnson CA, Mehdi SQ, Maher ER. Identification of
novel TYR and TYRP1 mutations in oculocutaneous albinism. Clin Genet 2005;68(2):182-184. [CrossRef] [PubMed]
8 Kaul H,
Riazuddin SA, Shahid M, Kousar S, Butt NH, Zafar AU, Khan SN, Husnain T, Akram
J, Hejtmancik JF, Riazuddin S. Autosomal recessive congenital cataract linked
to EPHA2 in a consanguineous Pakistani family. Mol Vis 2010;16:511-517. [PMC free article]
[PubMed]
9 Kaul H,
Riazuddin SA, Qazi ZA, Nasir IA, Zafar AU, Khan SN, Husnain T, Akram J,
Hejtmancik JF, Riazuddin S. Ectopialentis in a consanguineous pakistani family
and a novel locus on chromosome 8q. Arch
Ophthalmol 2010;128(8):1046-1049. [CrossRef] [PubMed] [PMC free article]
10 Lathrop GM,
Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in
humans. Proc Natl Acad Sci 1984;81(11):3443-3446.
[CrossRef]
11 Simeonov
DR, Wang X, Wang C, Sergeev Y, Dolinska M, Bower M, Fischer R, Winer D,
Dubrovsky G, Balog JZ, Huizing M, Hart R, Zein WM, Gahl WA, Brooks BP, Adams
DR. DNA variations in oculocutaneous albinism: an updated mutation list and
current outstanding issues in molecular diagnostics. Hum Mutat 2013;34(6):827-835. [CrossRef] [PubMed] [PMC free article]
12 Shah SA, Din SU,
Raheem N, Daud S, Mubeen J, Nadeem A, Tayyab M, Baloch DM, Babar ME, Ahmad J.
Identification of a novel mutation (p.Ile198Thr) in gene TYR in a Pakistani
family with nonsyndromicoculocutaneous albinism. Clin Exp Dermatol 2014;39(5):646-648.
[CrossRef] [PubMed]
[Top]