INTRODUCTION
Congenital cataracts are a significant cause of visual damage in children, resulting in approximately 10%of childhood blindness worldwide, with a prevalence of 0.6 to 6.0 in 10000 live births[1-2]. The forms of inherited nonsyndromic cataracts are autosomal dominant (AD) inheritance,autosomal ressive (AR) inheritance and X-linked inheritance,of which the AD form is the majority[3-5]. So far, over 39 independent loci and genes have been reported to be associated with inherited cataracts. These genes encode for crystallin,gap junctions, lens major intrinsic proteins, heat shock transcription factor-4, beadedfilament structural protein-2, and other proteins[6]. Closely packed crystallins are crucial for lens transparency and vision healthy. The major members of the crystallins are α-crystallin, β-crystallin and γ-crystallin. In the crystallin superfamily, α-crystallin are described as molecular chaperones related to the small heat shock proteins, and βand γ-crystallins are detected only in the eye and mainly in the ocular lens and found to share some common characteristics,such as a common core structure comprising Greek key motifs[7].
In the present study, we performed direct DNA sequencing of all exons and flanking intronic sequences of candidate genes associated with autosomal dominant congenital cataract(ADCC). A splice site mutation of c.30-2 A>G located two base pairs (bp) before the 5' donor splice site in exon 2 of βA1/A3-crystallin gene (CRYBA3/A1 gene) was identified. The CRYBA3/A1 gene, which is located in 17q11.2 and consists of six exons, is an important member of the β-crystallin family, encoding βA3-crystallin and βA1-crystallin[8]. To our knowledge, the previous four types of splice site mutations reported to be associated with ADCC in CRYBA3/A1 gene are all in the exon 3-intron 3 junction region[9-12]. Thus, we reported a novel mutation, CRYBA3/A1 gene c.30-2 A>G, associated with ADCC.
MATERIALS AND METHODS
Family Ascertainment and Genomic DNA Preparation A four-generation Chinese pedigree from Henan Province with a history of ADCC were recruited from the Eye Center of the Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang Province, China. Written informed consent adhered to the tenets of the Declaration of Helsinki, and this study was approved by the Zhejiang University Institutional Review Board. Family history and the history of cataract extraction were recorded. We collected 5 mL of venous blood from all individuals (three affected and one unaffected) from the family who took part in the study for genomic DNA extraction using the Simgen Blood DNA mini kit (Simgen, Hangzhou, China).
Mutation Identification All coding exons and exon-intron junctions of all congenital cataract-associated genes, namely CRYAA, CRYAB, CRYBA3/A1, CRYBB1, CRYBB2, CRYGC,CRYGD, GJA3, GJA8, and MIP were amplified by polymerase chain reaction (PCR) using specific primers and conditions,as described by Yu et al[13] in our previous study. The PCR products were then sequenced using the BigDye Terminator Cycle sequencing kit V3.1 (Applied Biosystems, Foster City,CA, USA) according to the manufacturer's directions and analyzed using Chromas 1.62. The consequent results werefinally compared with the National Center for Biotechnology Information (NCBI) reference sequences.
Single-strand Conformational Polymorphism Analysis and Bioinformatics Analysis PCR products were purified and digested using restriction endonuclease of BspEI (New England Biolabs, Inc.). The products were then separated on 1% agarose gels and stained with ethidium bromide. The presence of a mobility shift was determined by electrophoresis.In order to predict the affects of the mutation to the protein,several kinds of bioinformatics analysis including Human Splicing Finder Matrices and MaxEnt tools were conducted to analyze the differences in the splice patterns between the wild-type sequence and the mutated sequence. In addition, the Annovar software, as described by Taylor et al[14] and Geller et al[15] also estimated the pathogenicity to be the result of the A to G transition.
RESULTS
Clinical Evaluations Cataracts exhibited AD inheritance pattern in the four-generation pedigree (Figure 1). The proband(IV:1), a 17-year-old man, had undergone bilateral cataract surgery in the First Affiliated Hospital of Zhengzhou Medicine University when he was 7 years old, but the specific slit lamp photography was not provided when we began our study.Two other affected family members (III:1 and IV:2) also had a history of bilateral cataract surgery. There was no family history of other ocular or systemic abnormalities.
Gene Sequencing Direct sequencing revealed a heterozygous change, A to G, at position c.30-2 of the CRYBA3/A1 gene,which leads to a base change in the exon-intron splice site(Figure 2). This junction site is highly conserved across various species, as determined from the NCBI database. This mutation co-segregated with all affected individuals in the family and was not found in any of the unaffected family members or in the 100 unrelated normal controls.
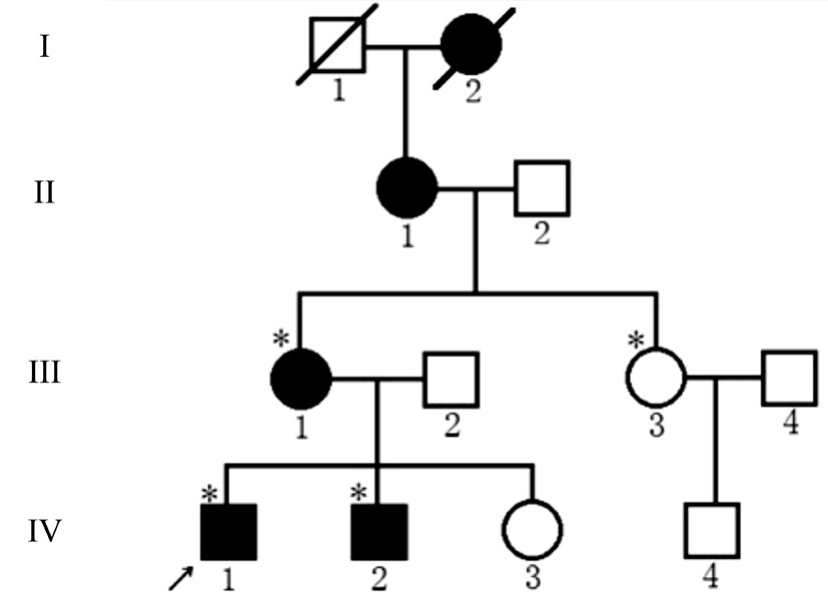
Figure 1 Pedigree of the four-generation family with ADCC Squares and circles indicate males and females, respectively. Black symbols indicate affected individuals, and open symbols indicate unaffected individuals. The diagonal line indicates a deceased family member, and asterisks indicate family members who participated in this study. The black arrow indicates the proband.
Single-strand Conformational Polymorphism Analysis A 275-bp fragment was PCR amplified from the DNA of each family member using the primers fl anking exon 2 of CRYBA3/A1 gene. The mutated sequence created a recognition site for the enzyme BspEI within this amplicon. When the amplicon from the five affected individuals was digested, it created a 170-bp and a 105-bp product, while the amplicons from the unaffected individuals were uncleaved 275-bp products.Sequencing confirmed the presence of c.30-2 A>G in the exon–intron junction region of CRYBA3/A1 gene in the affected individuals. Furthermore, none of the DNA samples from 100 unrelated normal controls showed this variation or were found to have the BspEI site. All related results are shown in Figure 3.
Bioinformatics Analysis The potential impact of the splice site mutation identified in this study was assessed using bioinformatic tools. The mutation was predicted to have a high risk of leading to a broken site and subsequently resulting in erroneous mature mRNA constitution according to the online Human Splicing Finder Matrices and MaxEnt tools, with the variation in the wild-type sequence estimated to be 89.89,and variation for the mutated sequence being 60.94 (ΔCV:-32.21%). In addition, the computer software Annovar gave a result of highly damaging, with a CADD score of 22.5,a MutationTaster score of 1.0, a GERP score of 5.72 and a phyloP score of 6.383, which was consisted to be potentially deleterious.
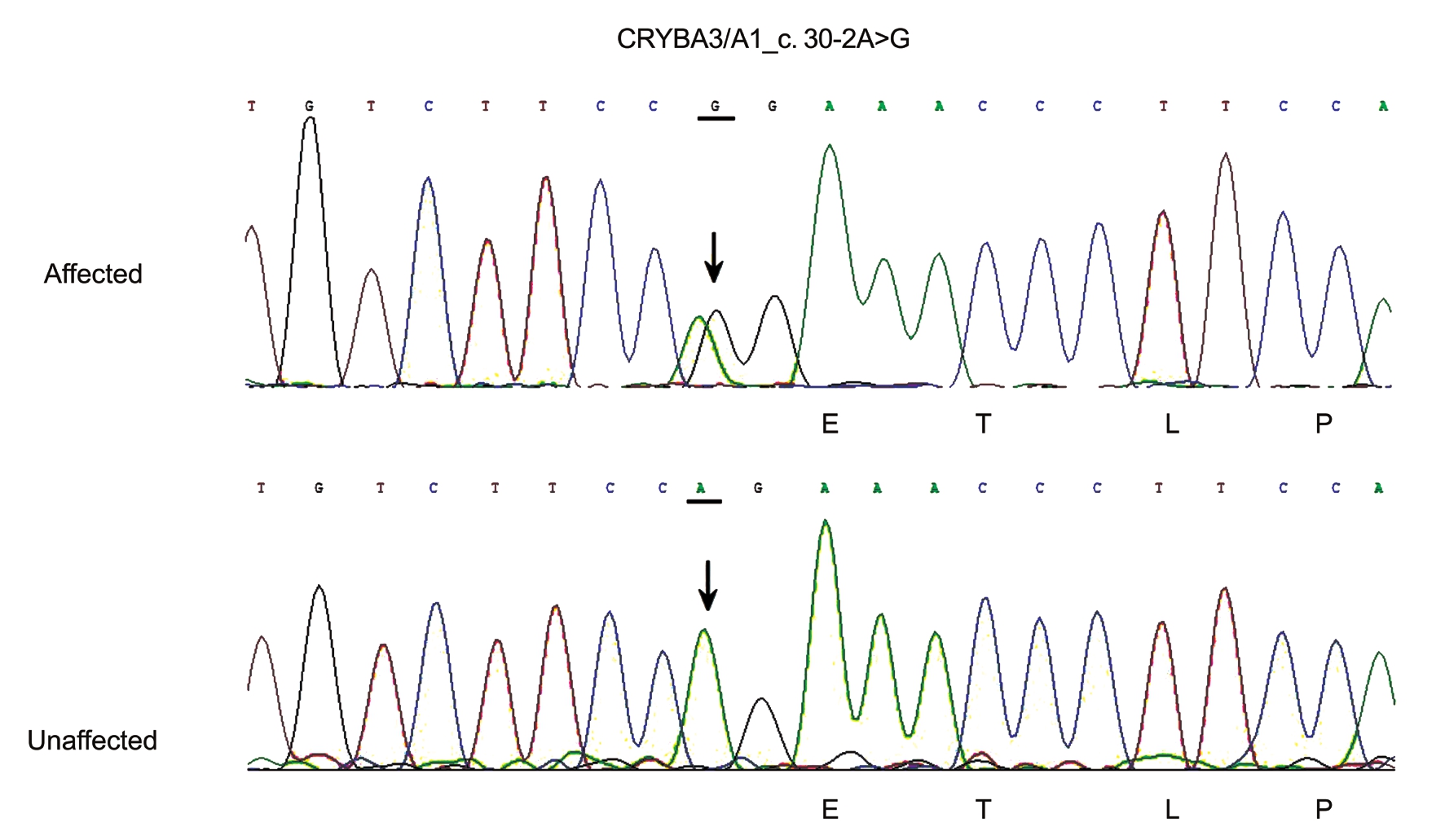
Figure 2 Partial nucleotide sequence of CRYBA3/A1 gene in this family A heterozygous A-to-G conversion was detected in all of the affected family members, resulting in a base change at the second position before the 5’ end of exon 2. The A-to-G conversion was not found in unaffected family members or 100 control subjects.

Figure 3 SSLP analysis of the mutation region in CRYBA3/A1 gene The fragment was cleaved to a 170-bp and a 105-bp fragment in the three affected individuals, whereas the product was a 275-bp fragment in the unaffected family member.
DISCUSSION
In this study, a novel splice site mutation (c.30-2 A>G) was identified in the CRYBA3/A1 gene in a Chinese pedigree affected by ADCC. The observed mutation was then evaluated within the available family members with single-strand conformational polymorphism (SSCP) analysis. Both direct sequencing of PCR products and SSCP analysis show the presence of the mutation. This novel base substitution was also confirmed to be absent in 200 chromosomes of the 100 unrelated normal controls, indicating that the mutation cosegregated within this family.
Abnormalities in lens crystallins were regarded as fundamental factors in formation of cataracts by failing to establish and maintain lens transparency[16-17], since the β- and γ-crystallins tend to be found primarily in the lens nucleus[18-19]. To our knowledge, there are seven protein regions in βA1/A3-crystallin: four twisted β-pleated sheets named Greek key motifs[7], one connecting peptide, and NH2- and COOH-terminal extensions. The first two exons encode the NH2-terminal arm, and exons 3, 4, 5 and 6 encode the Greek key motifs, which are organized into two domains[20]. In previous research, four types of splice site mutations in the CRYBA3/A1 gene are reported to be related to ADCC: IVS3+1 G>A[9],IVS3+1 G>C[10], IVS3+1 G>T[11], and IVS3+2 T>G[12]. Almost all of these splice site mutations identified previously were revealed to in fl uence the exon 3-intron 3 junction region and be responsible for improperly folded crystallins, which were unstable and accelerated the precipitation of other mutant proteins. In addition, they might also interfere with remaining β-crystallins[9]. Ultimately they led to opacification in the lens and formation of cataracts[21]. In the intron 1-exon 2 junction region, no splice site mutation has been previously reported.So far, few studies concerning the precise mechanisms of mature mRNA splicing around this region have been reported.Since this region was reported to be relevant to the structure of NH2-terminal arm[20], we could preliminarily consider that this alteration in protein structure might further influence the protein properties, such as solubility, and lead to protein denaturation.
Bioinformatic analysis were usually conducted in previous studies to study the functional properties of a mutation.In our study, the A at position c.30-2 is considered highly conserved. The presence of exon-intron splice sites was evaluated through parameters of variation (for the wild-type sequence and the mutated sequence, respectively) and the CV differences (ΔCV) using Human Splicing Finder Matrices and MaxEnt tools. Significant splice sites were not identified in the mutated sequence, according to the results obtained, showing a structural change of the sequence. This suggested that the A>G mutation at this position was tested for causation of the broken splice site, which has direct in fl uence on the splicing of the mature mRNA and results in abnormalities. Meanwhile,variants were annotated after retrieving relevant information from several public databases in order to obtain information from ExonicFunc, AAChange, SIFT, PolyPhen-2, LRT,MutationTaster, FATHMM, RadialSVM, LR, and VEST3 scores, CADD, GERP++, phyloP, SiPhy, cosmic70, ExAC,and OCT, using the Annovar software. Thus the mutation was predicted to be damaging and the pathogenicity was verified once again to be the result of the A to G transition, leading us to consider the conceivable results of splicing. Because this site was predicted to be unable to serve as a splice site, the outcome could be one of two kinds of abnormalities: intron 1 could remain as pre-mRNA with recruitment and activation of a cryptic splice site (a termination codon) or could result in mature mRNA consisting of only exon 1 and several mRNA isoforms of exons 3, 4 and 5. Either abnormality would produce truncated mRNA that would cause an unstable and dysfunctional protein and could possibly interfere with other appropriate associations of β-crystallin. Finally, cataracts could result from each of these abnormalities.
There are certainly limitations in this study. The precise mechanism of lens opacification as a result of this mutation should be studied further. Moreover, due to the history of cataract extraction of the proband 10y ago in another hospital,the specific slit-lamp photography was unavailable. This left us uncertain whether cataracts caused by the same CRYBA3/A1 gene mutation could be considered gene pleiotropy among affected individuals in this single pedigree in this study.This possibility remains to be further investigated. However,although there is no clear genotype–phenotype relationship in this pedigree, it is evident that the c.30-2 A>G mutation in CRYBA3/A1 gene causes aberrant splicing of the mature mRNA and is likely responsible for the cataracts in this Chinese family.
In conlusion, wefirstly reported a novel splice site mutation,c.30-2 A>G mutation of CRYBA3/A1 gene associated with ADCC. Our result broadens the genetic and mutation spectrum of ADCC. Further functional studies are needed to explore the potential mechanisms for the cataract formation.
REFERENCES
1 Ionides A, Francis P, Berry V, Mackay D, Bhattacharya S, Shiels A,Moore A. Clinical and genetic heterogeneity in autosomal dominant cataract. Br J Ophthalmol 1999;83(7):802-808.
2 Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol 2004;49(3):300-315.
3 Shiels A, Hejtmancik JF. Genetics of human cataract. Clin Genet 2013;84(2):120-127.
4 Rahi JS, Dezateux C. Congenital and infantile cataract in the United Kingdom: underlying or associated factors. British Congenital Cataract Interest Group. Invest Ophthalmol Vis Sci 2000;41(8):2108-2114.
5 Wirth MG, Russell-Eggitt IM, Craig JE, Elder JE, Mackey DA.Aetiology of congenital and paediatric cataract in an Australian population. Br J Ophthalmol 2002;86(7):782-786.
6 Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol 2008;19(2):134-149.
7 Hogg D, Tsui LC, Gorin M, Breitman ML. Characterization of the human beta-crystallin gene hu beta a3/a1 reveals ancestral relationships among the beta gamma-crystallin superfamily. J Biol Chem 1986;261(26):12420-12427.
8 Graw J, Jung M, Loster J, Klopp N, Soewarto D, Fella C, Fuchs H, Reis A, Wolf E, Balling R, Hrabe de Angelis M. Mutation in the betaa3/a1-crystallin encoding gene cryba1 causes a dominant cataract in the mouse.Genomics 1999;62(1):67-73.
9 Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaa3/a1-crystallin gene. Mol Vis 1998;4:21.
10 Bateman JB, Geyer DD, Flodman P, Johannes M, Sikela J, Walter N, Moreira AT, Clancy K, Spence MA. A new betaa1-crystallin splice junction mutation in autosomal dominant cataract. Invest Ophthalmol Vis Sci 2000;41(11):3278-3285.
11 Yang Z, Li Q, Ma Z, Guo Y, Zhu S, Ma X. A G-T splice site mutation of cryba1/a3 associated with autosomal dominant suture cataracts in a Chinese family. Mol Vis 2011;17:2065-2071.
12 Yang Z, Su D, Li Q, Yang F, Ma Z, Zhu S, Ma X. A novel T-G splice site mutation of cryba1/a3 associated with autosomal dominant nuclear cataracts in a Chinese family. Mol Vis 2012;18:1283-1288.
13 Yu Y, Li J, Xu J, Wang Q, Yu Y, Yao K. Congenital polymorphic cataract associated with a g to a splice site mutation in the human beta-crystallin gene crybetaa3/a1. Mol Vis 2012;18:2213-2220.
14 Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet 2015;47(7):717-726.
15 Geller F, Feenstra B, Carstensen L, et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat Genet 2014;46(9):957-963.
16 Hejtmancik JF. The genetics of cataract: Our vision becomes clearer.Am J Hum Genet 1998;62(3):520-525.
17 Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE,Elder JE, Dickinson JL, Sale MM. Investigation of crystallin genes in familial cataract, and report of two disease associated mutations. Br J Ophthalmol 2004;88(1):79-83.
18 Piatigorsky J. Lens crystallins and their gene families. Cell 1984;38(3):620-621.
19 Aarts HJ, Lubsen NH, Schoenmakers JG. Crystallin gene expression during rat lens development. Eur J Biochem 1989;183(1):31-36.
20 Wistow G, Turnell B, Summers L, Slingsby C, Moss D, Miller L,Lindley P, Blundell T. X-ray analysis of the eye lens protein gamma-II crystallin at 1.9 A resolution. J Mol Biol 1983;170(1):175-202.
21 Graw J. Genetics of crystallins: Cataract and beyond. Exp Eye Res 2009;88(2):173-189.