INTRODUCTION
Myopia is a common eye disease and a significant public health issue. Simple myopia can be corrected with spectacles or contact lenses, whereas high (pathological)myopia is one of the main causes leading to low vision or blindness. Pathological myopia is defined as a refractive error of at least −6.00 diopters (D) with pathological changes including temporal crescent, pigment epithelial thinning,leopard retina, Fuch’s macula, and retina-choroidal atrophy.Epidemiological evidence suggests increasing prevalence of myopia, especially in Asian populations[1-2], causing profound economic cost to the society. Therefore, it is important to identify the etiology of high myopia.
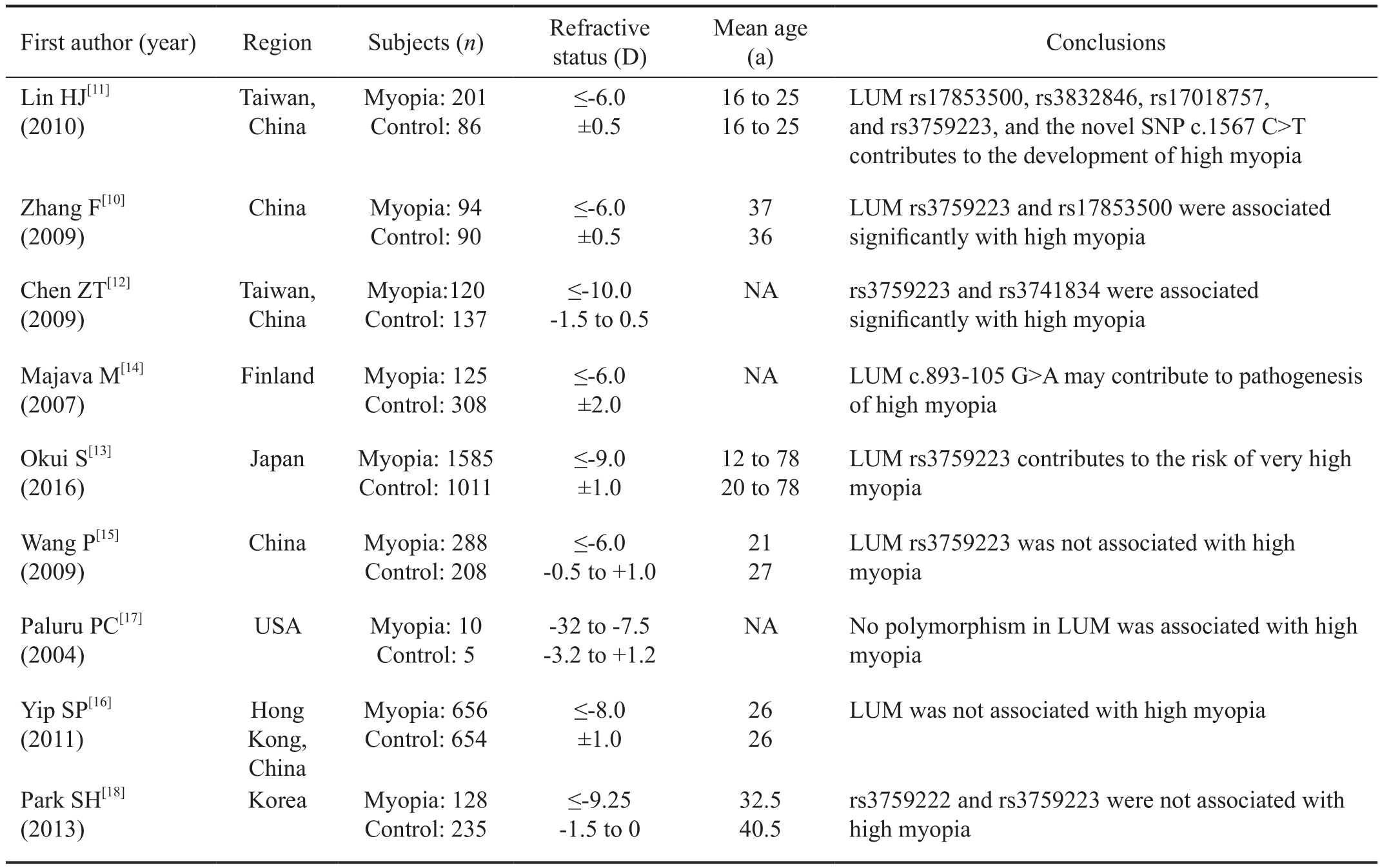
Active remodeling mechanism of the sclera is an important process during the pathogenesis of myopia. Scleral remodeling involves reduced production of collagen and proteoglycans and increased collagen degradation. The sclera contains a collagenrich extracellular matrix that undergoes significant biochemical and biomechanical remodeling during the development of myopia[3-5]. Alteration in the expression levels of extracellular matrix components may affect scleral morphology, as in myopia. Lumican (LUM), a keratan sulfate proteoglycan belonging to the small leucine-rich proteoglycan (SLRP)family, is one of the major extracellular matrix components of the sclera. LUM-deficient mice displayed corneal opacity,skin fragility, abnormally large collagen fibril diameters, and disorganized inter fibrillar spacing[6]. This role implicated LUM could be responsible for the control of collagen fibrillogenesis.And Chakravarti et al’s[7] study further showed that mice deficient in LUM and fibromodulin manifested certain features of high myopia. LUM knockdown zebrafish caused scleral thinning and increased size of scleral coats[8]. LUM mRNA level was significantly down-regulated during the development of lens-induced myopia[9]. It’s suggested that altered expression of LUM may contribute to myopia. Increasing studies have showed that single nucleotide polymorphisms (SNPs) in the LUM gene were associated with high myopia[10-13]. Majava et al[14] found that a novel SNP c.893-105G>A may contribute to pathogenesis of high myopia. However, some other studies suggested that polymorphisms in LUM gene were not associated with high myopia[15-18]. Case-control studies about the relationship between LUM polymorphisms and high myopia were summarized in Table 1. Therefore, we want toinvestigate whether LUM gene polymorphisms are correlated with high myopia in the Southern Chinese population.LUM gene was extracted from the blood of 95 patients with high myopia and 95 individuals in the control group, and sequenced the polymorphisms of LUM coding region by direct sequencing.
Table 1 Summary of case-control studies about the relationship between LUM polymorphisms and high myopia
SUBJECTS AND METHODS
Patients and Study Design From January to December 2012,we recruited 500 volunteers and examined their refractive status in detail at the First Affiliated Hospital of Jinan University.All of the participants were unrelated and of Chinese Han Nationality. The eligible volunteers were divided into the pathological myopia group and control group with 95 members in each group. LUM gene was extracted from the blood of two groups, and sequenced the polymorphisms of LUM coding region by direct sequencing. This research followed the Declaration of Helsinki and conformed to the principles of medical ethics. For each subject, the study protocol and procedure were fully explained, and written consent was obtained. The experiment was authorized by the First Affiliated Hospital of Jinan University Ethics Committee.
Subject Enrollment The two group were given detailed ophthalmic examinations, including uncorrected visual acuity and best corrected visual acuity, subjective refraction, ocular movements, slit-lamp examination (Topcon, Japan), noncontact intraocular pressure (IOP; Topcon, Japan), optical coherence tomography (Zeiss Humphrey, Germany) and fundus color photography (Topcon, Japan). Anterior chamber depth (ACD) and axial length (AXL) is measured by IOL master (Zeiss Humphrey, Germany). In accordance with pathological myopia diagnostic criteria, clinical presentation of the disease included: 1) binocular myopic spherical equivalent(SE) >6.0 D; 2) AXL >26 mm; 3) with one of the following ocular fundus changes: leopard retina, comus, lacquer crack,macular hemorrhage, Fuchs spot, posterior staphyloma,peripheral chorioretinal lesions and retinal detachment.Inclusion criteria for the control group included: 1) binocular SE between -0.50 D to +0.50 D; 2) AXL <24 mm; 3) no family history of pathological myopia. Participants with eye disease,history of intraocular surgery, cataract, glaucoma, retinal disorders, or laser treatment were excluded.
Lumican Gene Sequencing The genomic DNA was extracted from 5 to 10 mL of venous blood from all participants.DNA was purified from lymphocyte pellets according to standard procedures using a Puregene DNA isolation kit (Tiangen, Beijing, China). The primer sequences used in the polymerase chain reaction (PCR) are as follows: LUM-1F: GCACGTGTACGCAATCTAACC;LUM-1R: GTTGTGCAGCCCAGGTATTT; LUM-2F: ACTGTGCCATTTTGGTAGCC; LUM-2R:GGCCTGCCTTTCATCTTTTC. The PCR was performed in a 30 μL reaction volume containing 3 μL 10× rTaq buffer(Tiangen, Beijing, China), 200 μmol/L of dNTP, 0.2 μmol/L of each primer, 1.0 unit of rTaq DNA polymerase (Tiangen,Beijing, China), and 80 ng of genomic DNA. The conditions,after initial denaturation at 95℃ for 5min, were 30 cycles of 30s at 95℃, 30s at 55℃, and 45s at 72℃, followed by final extension at 72℃ for 5min. The PCR products remained at 12℃ and were purified using a Multi Screen-PCR plate(Millipore, Boston, USA). The purified PCR products were bidirectionally sequenced using the ABI3730XLDNA sequencer (Applied Biosystems, Foster, USA).
Statistical Analysis Characteristics between the two groups were tested using independent-samples t-test or Levene’s test for equality of variances. Genetic analysis and Hardy-Weinberg equilibrium were analyzed through Chi-square test or Fisher’s exact test and P-value were calculated by SPSS 16.A P-value <0.05 was considered significant.
RESULTS
Patient Characteristics The study group was comprised of 95 patients with high myopia and the control group consisted of 95 individuals with normal vision. The patients enrolled in the study group were with age 18-30y, male:female ratio was 1.1:1.0, mean ACD 3.46 mm, mean cornea diopter (CD) 43.4 D,and mean IOP 16 mm Hg. The volunteers in the control group were with age 18-30y, male: female ratio was 0.7:1.0, mean ACD 2.99 mm, mean CD 43.2 D, and mean IOP 15.7 mm Hg (Table 2). There was no significant difference between the control and study groups in age, gender, CD, ACD, and IOP.
Hardy-Weinberg Equilibrium Analysis SNPs in the coding region of LUM gene were determined for investigating whether LUM gene polymorphisms correlate with high myopia.Direct-sequencing analysis found three SNPs of the LUM gene in coding region, which were LUM c.32 (rs577456426),LUM c.507 (rs17853500) and LUM c.849 (rs181915277).The genotype distributions were obtained by direct counting and allele frequencies were calculated subsequently. The genotype distributions of the three SNPs between the patients and control subjects were consistent with Hardy-Weinberg equilibrium (Table 3).
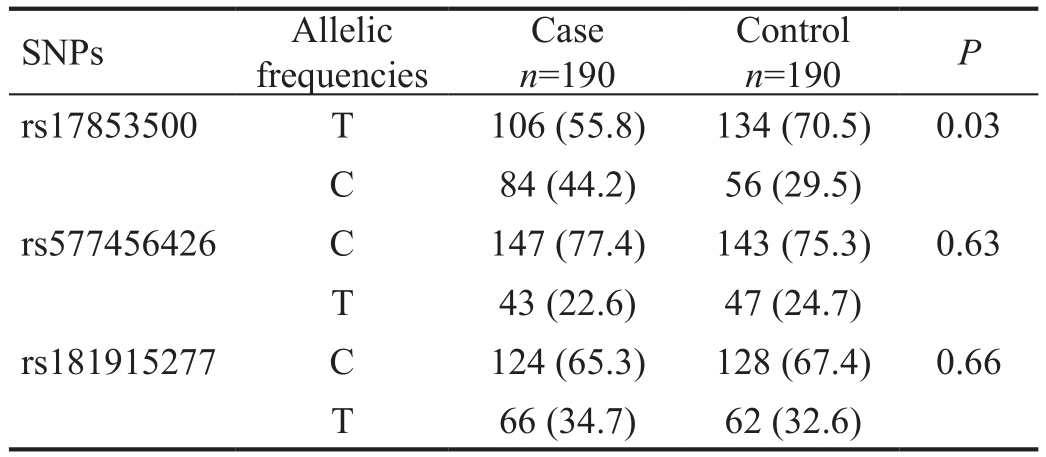
Lumican Gene Sequencing Analysis This study detected three polymorphisms of the LUM gene, which are LUM c.32(rs577456426), LUM c.507 (rs17853500) and LUM c.849(rs181915277). The genotype distributions and allelic frequencies of the three SNPs between the high myopia and control groups were shown in Tables 3 and 4. For the polymorphism rs17853500, the genotype frequencies of T/T:T/C:C/C were 26.3%:58.9%:14.7% respectively, in the high myopia group and 50.5%:40.0%:9.5% respectively,in the control group. The allelic frequencies of T:C was 55.8%:44.2% in the high myopia group and 70.5%:29.5%in the control group. There was a significant relationship between rs17853500 of LUM gene and high myopia (P<0.05).However, there were no significant differences in genotypedistributions and allelic frequencies of rs577456426 and rs181915277 between the two groups (P>0.05) (Tables 4, 5).
Table 2 Characteristics of the high myopia patients and control subjects mean±SD
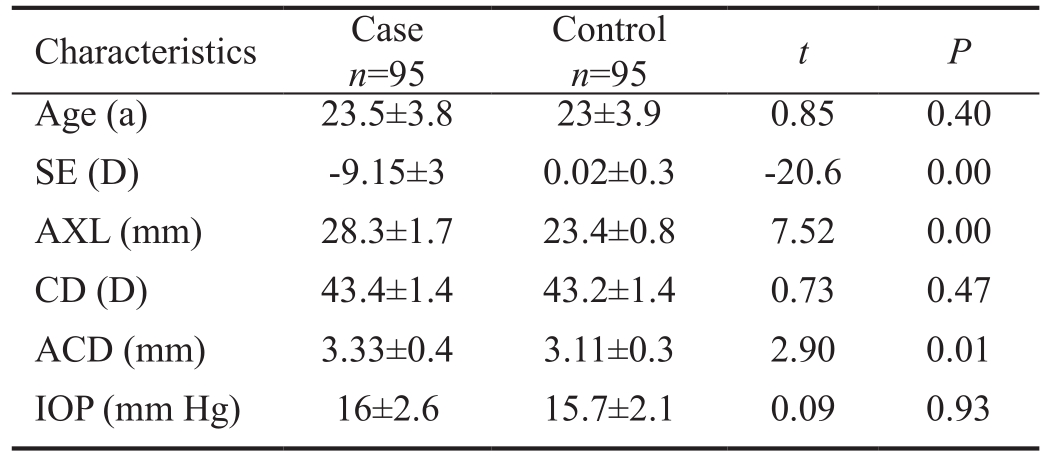
SE: Spherical equivalent; AXL: Axial length; CD: Cornea diopter;ACD: Anterior chamber depth; IOP: Intraocular pressure; SD:Standard deviation.
DISCUSSION
As a member of the SLRP family, LUM is widely expressed in fibrous connective tissue, participating in the regulation of stromal collagen fibrils assembly, corneal transparency, scleral morphology, skin elasticity and so on[19-21]. LUM is located at 12q21-q23 (MYP3), a genetic locus often associated with high myopia[22]. Gene knockout studies[6-8] suggested that LUM was a potential pathological myopia gene. The study explored the association of LUM polymorphisms and high myopia in the Southern Chinese population.
This genetic association study revealed that there was a significant difference in the polymorphism rs17853500 of LUM between high myopia patients and normal controls,whereas no significant difference was detected in the other two polymorphisms (rs577456426 and rs181915277). These results indicated that LUM rs17853500 at the coding region of might be associated with increased susceptibility to high myopia. Our results are consistent with Zhang et al’s[10] and Lin et al’s[11] studies. Zhang et al[10] revealed that SNPs rs17853500 and rs3759223 in LUM were associated significantly with pathological myopia in Northern Han Chinese, and they also found that there was no difference in the rate of mutation between pedigree and sporadic group. Lin et al’s[11] study revealed that haplotype distributions of LUM polymorphisms(rs17853500, rs3832846, rs17018757 and rs3759223)contributed to the pathogenesis of high myopia in Taiwan-born Han Chinese. They sequenced all three exons, intron-exon boundaries, and promoter regions of LUM, while we detected the SNPs of LUM gene in coding region. However, Paluru et al[17] and Park et al [18] did not support an association of LUM polymorphisms with high myopia in the USA and Korea respectively. The contradict result from these studies might be explained by racial and region differences. Moreover, these studies used different genotyping methods. They analyzed the SNPs genotyping by restriction fragment length polymorphism(RFLP), mass spectrometry or DNA pooling. We detected theSNPs of LUM encoding region by direct sequencing, which was known as the gold standard for detecting polymorphisms.Nevertheless, direct sequencing requires the PCR ampli fi cation of each site, and is not suitable for the screening of the whole genome.
Table 3 Hardy-Weinberg equilibrium analysis of the three SNPs in the high myopia patients and control subjects
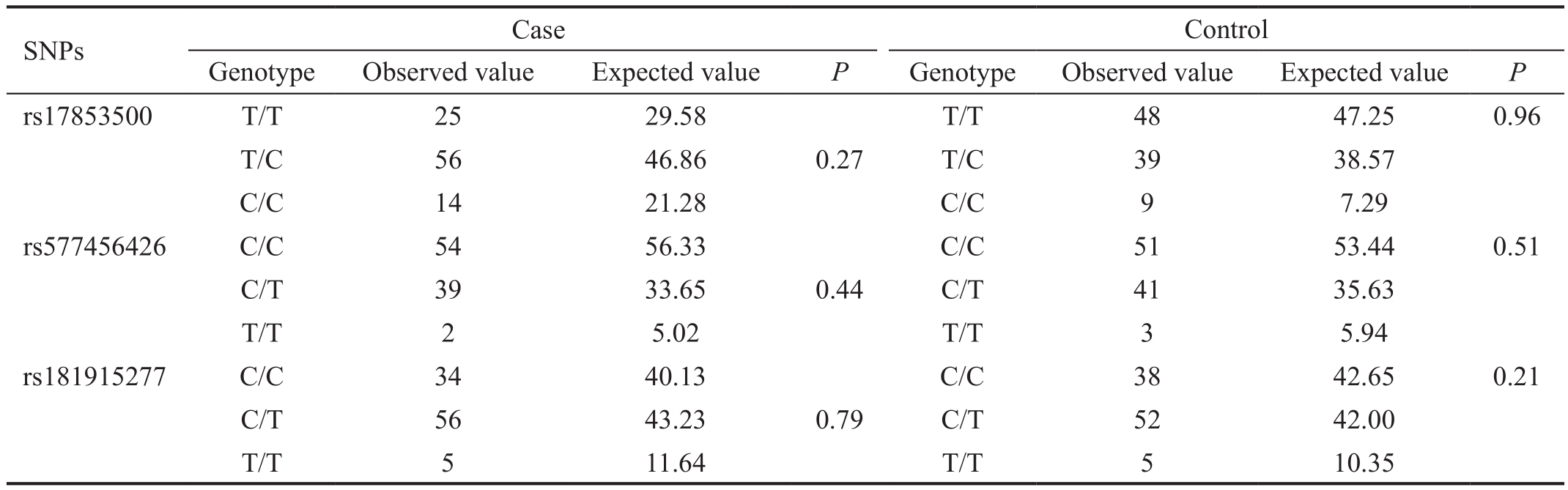
Table 4 Genotype distributions of the three SNPs in the high myopia patients and control subjects n (%)
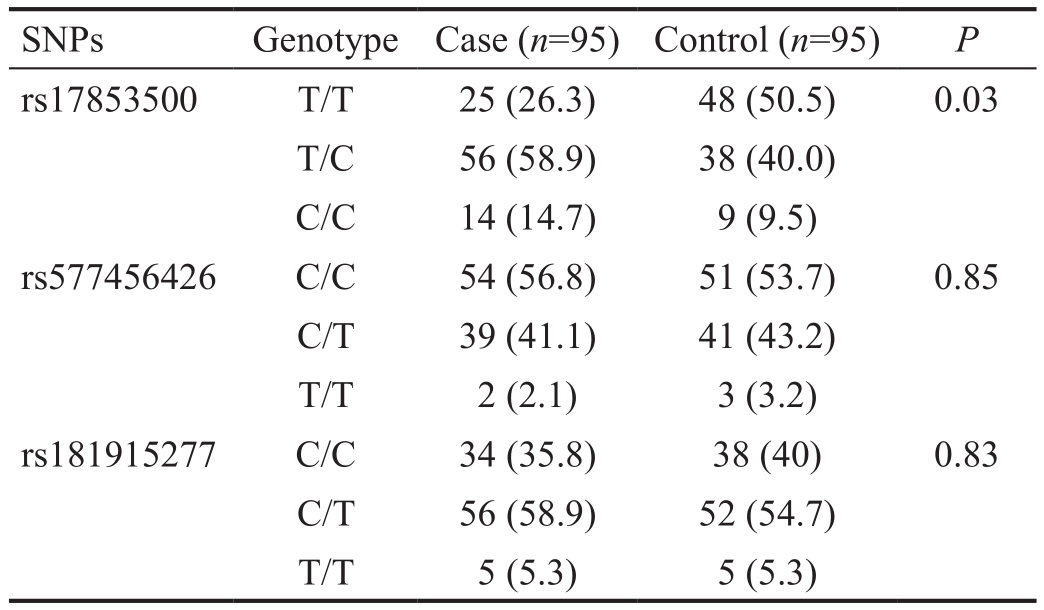
Table 5 Allelic frequencies of the three SNPs in the high myopia patients and control subjects’ eyes n (%)
This study investigated the relationship between LUM polymorphisms and high myopia by a case-control study,which is an effective, economic and simple method to study the etiology and risk factors of the disease. It can be applied to the evaluation of disease risk factors, monitoring of drug efficacy[23], evaluation of vaccine immunological effect[24], and outbreak investigation[25], etc. However, the results of casecontrol studies were influenced by the researcher’s design flaws, such as the source and selection of cases and controls,the size of the sample, the measurement and regulation of the relevant factors, and so on. Wang et al[15] did not support an association of LUM rs3759223 with high myopia in China. In their study, they just detected one SNP rs3759223 of LUM by RFLP analysis, whereas we analyzed the SNPs of LUM gene in coding region by direct sequencing. The control individuals from their study had bilateral refraction ranging from -0.5 to 1.0 D, whereas in our study, the control subjects had refractive errors between -0.50 and +0.5 D. In addition, the participants in their study were larger than that in our group. These different selection criteria might contribute to the discrepancy between the findings in the two studies. This present study has some limitations, including regional limitations of population selection and small sample size. Further studies can be carried out in a multicenter study with a number of hospitals across the country, so as to increase the sample size and reduce the selection bias.
In conclusion, our findings demonstrated that rs17853500 polymorphism was correlated with high myopia. LUM might play an important role in regulating scleral extraccllular matrix interactions, thus affecting the development of myopia. Further research is required to con firm whether LUM is the virulence gene of high myopia, and the role of LUM gene mutation in the pathogenesis of high myopia.
ACKNOWLEDGEMENTS
Foundations: Supported by Natural Science Foundation of Guangdong Province, China (No.2015A030310158;No.2014A030313359); the Science and Technology Planning Project of Guangdong Province, China (No.2015B020226003);the Scientific and Cultivation Foundation of the First Affiliated Hospital of Jinan University (No.2015201).
Conflicts of Interest: Wang GF, None; Ji QS, None; Qi B,None; Yu GC, None; Liu L, None; Zhong JX, None.
REFERENCES
1 Rim TH, Kim SH, Lim KH, Choi M, Kim HY, Baek SH; Epidemiologic Survey Committee of the Korean Ophthalmological Society. Refractive Errors in Koreans: The Korea National Health and Nutrition Examination Survey 2008-2012. Korean J Ophthalmol 2016;30(3):214-224.
2 Wu LJ, You QS, Duan JL, Luo YX, Liu LJ, Li X, Gao Q, Zhu HP,He Y, Xu L, Jonas JB, Wang W, Guo XH. Prevalence and associated factors of myopia in high-school students in Beijing. PLoS One 2015;10(3):e0120764.
3 McBrien NA. Regulation of scleral metabolism in myopia and the role of transforming growth factor-beta. Exp Eye Res 2013;114(9):128-140.
4 Jonas JB, Xu L. Histological changes of high axial myopia. Eye (Lond)2014;28(2):113-117.
5 McBrien NA, Metlapally R, Jobling AI, Gentle A. Expression of collagen-binding integrin receptors in the mammalian sclera and their regulation during the development of myopia. Invest Ophthalmol Vis Sci 2006;47(11):4674-4682.
6 Chakravarti S, Magnuson T, Lass JH, Jepsen KJ, LaMantia C, Carrol H. Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican. J Cell Biol 1998;141(5):1277-1286.
7 Chakravarti S, Paul J, Robert L, Chervoneva I, Oldberg A, Birk DE.Ocular and scleral alterations in gene-targeted lumican-fibromodulin doublenull mice. Invest Ophthalmol Vis Sci 2003;44(6):2422-2432.
8 Yeh LK, Liu CY, Kao WW, Huang CJ, Hu FR, Chien CL, Wang IJ.Knockdown of zebra fi sh lumican gene (zlum) causes scleral thinning and increased size of scleral coats. J Biol Chem 2010;285(36):28141-28155.
9 Siegwart Jr JT, Strang CE. Selective modulation of scleral proteoglycan mRNA levels during minus lens compensation and recovery. Mol Vis 2007;13(10):1878-1886.
10 Zhang F, Zhu, T, Zhou Z, Wu Y, Li Y. Association of lumican gene with susceptibility to pathological myopia in the northern han ethnic Chinese. J Ophthalmol 2009;2009(7):514306.
11 Lin HJ, Kung YJ, Lin YJ, Sheu JJ, Chen BH, Lan YC, Lai CH,Hsu YA, Wan L, Tsai FJ. Association of the lumican gene functional 3'-UTR polymorphism with high myopia. Invest Ophthalmol Vis Sci 2010;51(1):96-102.
12 Chen ZT, Wang IJ, Shih YF, Lin LL. The association of haplotype at the lumican gene with high myopia susceptibility in Taiwanese patients.Ophthalmology 2009;116(10):1920-1927.
13 Okui S, Meguro A, Takeuchi M, Yamane T, Okada E, Iijima Y,Mizuki N. Analysis of the association between the LUM rs3759223 variant and high myopia in a Japanese population. Clin Ophthalmol 2016;10(10):2157-2163.
14 Majava M, Bishop PN, Hägg P, Scott PG, Rice A, Inglehearn C,Hammond CJ, Spector TD, Ala-Kokko L, Männikkö M. Novel mutations in the small leucine-rich repeat protein/proteoglycan (SLRP) genes in high myopia. Hum Mutat 2007;28(4):336-344.
15 Wang P, Li S, Xiao X, Jia X, Jiao X, Guo X, Zhang Q. High myopia is not associated with the SNPs in the TGIF, lumican, TGFB1, and HGF genes. Invest Ophthalmol Vis Sci 2009;50(4):1546-1551.
16 Yip SP, Leung KH, Ng PW, Fung WY, Sham PC, Yap MK. Evaluation of proteoglycan gene polymorphisms as risk factors in the genetic susceptibility to high myopia. Invest Ophthalmol Vis Sci 2011;52(9):6396-6403.
17 Paluru PC, Scavello GS, Ganter WR, Young TL. Exclusion of lumican and fibromodulin as candidate genes in MYP3 linked high grade myopia.Mol Vis 2004;10(11):917-922.
18 Park SH, Mok J, Joo CK. Absence of an association between lumican promoter variants and high myopia in the Korean population. Ophthalmic Genet 2013;34(1-2):43-47.
19 Stamov DR, Muller A, Wegrowski Y, Brezillon S, Franz CM.Quantitative analysis of type I collagen fibril regulation by lumican and decorin using AFM. J Struct Biol 2013;183(3):394-403.
20 Song Y, Zhang F, Zhao Y, Sun M, Tao J, Liang Y, Ma L, Yu Y, Wang J,Hao J. Enlargement of the axial length and altered ultrastructural features of the sclera in a mutant lumican transgenic mouse Model. PLoS One 2016;11(10):e0163165.
21 Chen S, Young MF, Chakravarti S, Birk DE. Interclass small leucinerich repeat proteoglycan interactions regulate collagen fi brillogenesis and corneal stromal assembly. Matrix Biol 2014;35:103-111.
22 Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS,Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet 1998;63(5):1419-1424.
23 Kus T, Aktas G, Alpak G, Kalender ME, Sevinc A, Kul S, Temizer M,Camci C. efficacy of venlafaxine for the relief of taxane and oxaliplatininduced acute neurotoxicity: a single-center retrospective case-control study. Support Care Cancer 2016;24(5):2085-2091.
24 Fletcher HA, Filali-Mouhim A, Nemes E, Hawkridge A, Keyser A,Njikan S, Hatherill M, Scriba TJ, Abel B, Kagina BM, Veldsman A,Agudelo NM, Kaplan G, Hussey GD, Sekaly RP, Hanekom WA, BCG study team. Human newborn bacille Calmette-Guérin vaccination and risk of tuberculosis disease: a case-control study. BMC Med 2016;14(16):76.
25 Makoni AC, Chemhuru M, Bangure D, Gombe NT, Tshimanga M.Rubella outbreak investigation, Gokwe North District, Midlands province,Zimbabwe, 2014 - a case control study. Pan Afr Med J 2015;22(22):60.