INTRODUCTION
M edicine is evolving rapidly into a genomic medicine discipline after successful sequencing of human and non-human genomes. As a result, traditional diagnostics,therapeutics and prognostics in patients will soon become outdated. In short, recent advances in genomics have led to identification of new genes and variants responsible for a host of inherited and inflammatory diseases in ophthalmology specialty. In a true sense, developments have made a huge impact in our understanding of genetic basis of structural and functional defects and disease mechanisms underlying eye diseases. For example, in past genetic causes of Leber congenital amaurosis (LCA); an inherited retinal dystrophy which leads to severe vision loss, and age-related macular degeneration (AMD); one of the most common causes of blindness were unknown. Today, more than 20 LCA genes have been identified, and now genetic testing can be performed in majority of cases. More importantly, gene-specific treatments have also been tested clinically. As far as AMD is concerned, it is a complex disease caused by a combination of genetics and environment[1]. Despite its complexity more than 40 loci have been accounted for 15% to 65% of AMD pathology. Thus, it appears that genetic underpinnings do play role in the causation and progression of eye ailments; a few of them are listed in Table 1 and briefly covered in this review. Hereditary and inflammatory diseases affect eyes in millions of people worldwide. Vision impairment is expected to rise as diseases that have measurable genetic components continue to grow[2]. There are about 4000 diseases/syndromes that affect humans and surprisingly one third involves eyes.Hence, it is important that improvement in knowledge of diseases will be useful to ophthalmologists and researchers equally to enrich abilities in providing accurate diagnosis,counselling, and treatments. It should be mentioned that ocular disorders with complex inheritance are responsible for most blindness. Therefore, developments in molecular genetics can change face of medicine in future. This has clear implications for physicians and their patients because these advancements will bring benefit to public. Within next few years, geneticists shall be able to complete a catalog of genes associated with eye diseases. Such progress will create possibilities, if not a mandate, for translational research in how best to apply discoveries for improving health. Driving these changes are advances in infrastructure, analytical methods, and training of workforce. This expansion will take us from a simple genetics paradigm wherein influence of individual genes on health will be paramount, to a real genomic medicine paradigm where effect of individual gene or variant will be considered together and in concert with environmental influence on one’s health outcome. As of today, genetic testing is still not performed for many conditions. This will change as we move forward to options that shall depend upon one’s own genotype. Similarly,identification of biomarkers in blood using ‘omics’ such as transcriptomics, epigenomics, proteomics, etc. will improve value of predictive tests aimed at understanding mechanism(s)of diseases. It is possible that such tests might also help stratify patients for suitable therapies thus enabling development of precision medicines (medicines that can enable disease prevention by delivering superior therapeutics by integrating clinical, multi-omics including epigenetics, environmental and behavioral information to understand biological basis of disease). It will lead to better selection of targets and identification of populations that will demonstrate improved outcomes to novel preventive and therapeutic approaches appropriately tailored to patients’ health outcomes[3]. Recently,gene therapy has advanced further leading to improvement in vision impairment thus giving hope because inherited diseases such as congenital cataracts, glaucoma, retinal degeneration,and optic atrophy affect quite many infants worldwide[4].Other pathologies that also exhibit genetic manifestations include AMD, Marfan syndrome (MFS), myopia, polypoidal choroidal vasculopathies (PCV), retinitis pigmentosa (RP),Stargardt’s disease (SD), and uveal melanoma (UM). Table 1 mentions a brief account of genes/variants that are involved in eye diseases. In adults, diseases such as glaucoma and AMD are leading causes of blindness across geographies and both show a strong genetic susceptibility. It is our belief that as breakthroughs continues to unfold in the form of innovative instrumentations along with novel bioinformatics pipelines,genomics of eye will also continue to advance. Approaches such as next-generation sequencing (NGS) are paving way for identification of newer mutations implicated in pathologies.These and other technologies such as genome-wise association studies (GWAS) or whole-exon sequencing (WES) have started helping us in using information in clinics for precise diagnosis,treatment response predications and related intervention(s).Our manuscript attempts to highlight ‘ongoing’ progress and future challenges encompassing field of investigative genomics as applied to the following diseases.
Table 1 Genes/gene variants linked with common human eye diseases/disorders

Age-related Macular Degeneration A leading cause of blindness AMD affects elderly people with an overall prevalence of about 8%. Since risk for AMD increases with age, its prevalence will continue to rise as population of older people increases. Globally, projected number of individuals with AMD in 2020 is 196 million and is expected to be 288 million by 2040[5]. It is characterized by degeneration of macula (which harbors fovea) resulting in loss of central vision[6]. Person with a normal vision sees grid lines without waves (not blurry) (Figure 1A), but someone with AMD sees grid lines as wavy or distorted (Figure 1B). While AMD etiology remains unclear, past advances have generated attractive patho-mechanistic hypotheses over the years including homocysteine (Hcy) mediated inflammation[7].Evidence suggests that age, smoking, and genetics are factors for susceptibility[8]. Many single nucleotide polymorphisms(SNPs) have been discovered in genes such as NOS2A which was found to increase risk especially SNP rs8072199 and with significant interaction along with smoking at SNP rs2248814.Stratification by genotypes demonstrated AMD and smoking in carriers of AA genotypes than AG or GG suggesting an interaction of AA with smoking[9]. Projects encompassing GWAS have revolutionized investigations with findings wherein variants were shown to confer increased likelihood of AMD. Results from eight variants in five genes showed a strong association to AMD for CFH rs1061170 CC, rs2274700 CC; for C2 rs9332739 CC/CG; for CFB rs641153 TT/CT; for HTRA1/LOC387715 rs10490924 TT; and for complement factor 3 (C3) rs2230199 GG. CFH confers increased risk to bilateral geographic atrophy whereas HTRA1/LOC387715 contributes more to bilaterality of choroidal neovascularization.Interestingly, C3 confers enhanced risk for geographic atrophy than choroidal neovascularization[10-15]. Advanced AMD has limited therapeutic options although both wet and dry AMD subtypes exhibit shared genetics. Recently a first genetic association specific to wet AMD near MMP-9 was discovered.Rare coding variants in CFH, CFI and TIMP3 suggested causal roles as did a splice variant in SLC16A8 supporting notion that rare coding variants can pinpoint genes within known genetic loci[16]. It is worth mentioning that AMD is different from other retinal conditions because risk for AMD involves a complex mix of genetics and lifestyles (Figure 2). However, genomics turned out to be useful in differencing and finding disease genes specific for AMD such as identification of CFH and its Y402H variant[17-19]. HTRA1 was also mapped by GWAS in etiology of AMD[20]. Therefore, it does bring home a point that genetic testing could help early detection, risk prediction and prognosis. Further, targeting high-risk individuals to reduce blindness through a robust surveillance and clinical interventions may help lessen overall AMD burden. Again,with the heritability of AMD estimated between 45% and 70%,it is important to rely upon genetic testing.

Figure 1 Worsening AMD or other macula related disorders Macula in these indications is damaged or degenerated thus making the grid lines (A) look wavy, blurred and distorted (B) in an Amsler Grid.
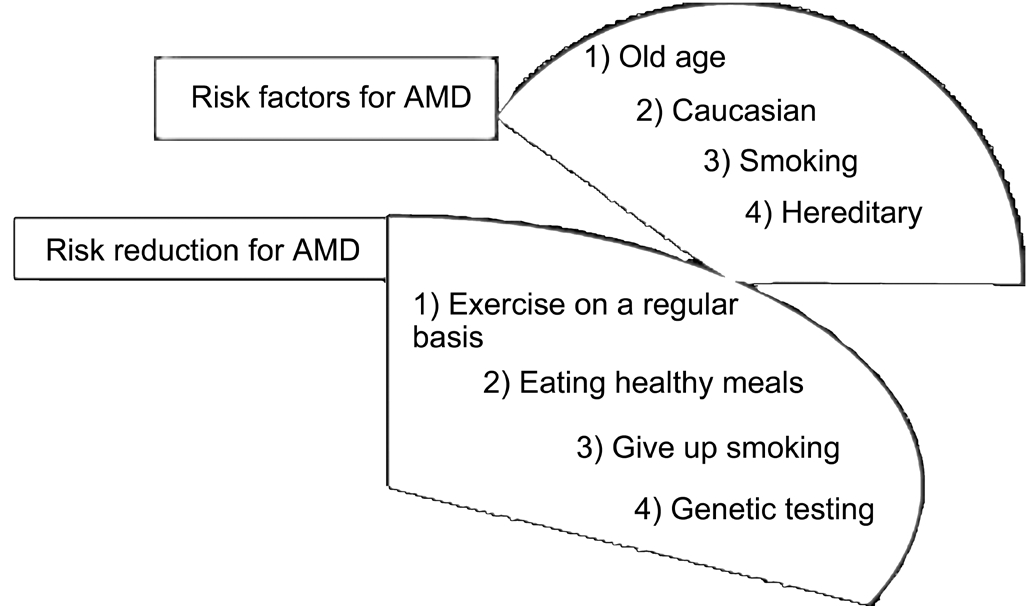
Figure 2 Prominent risk factors and their reduction in an AMD patient Suitable remedial action can in fact make things better for AMD susceptible individuals.
Cataract Cataract is a clouding of lens and congenital cataract is an important cause of blindness. It can result as a Mendelian inheritance. Up to 30% percent of cataracts are hereditary because of gamma crystallin genes. Clinical heterogeneity has been observed with gene mutations. Often, cataracts are related to aging. It can occur in both eyes, but does not spread from one eye to another. Transparency and high refractive index of lens are achieved by precise architecture of fiber cells along with homeostasis of lens proteins that are highly structured in terms of their concentrations, stabilities, and supramolecular organization. Their qualities maintain lens functioning. Genes encoding these components can determine lens health as congenital cataract is seen within first year of life, but juvenile cataract occurs during first decade. Pediatric cataract is heterogeneous in nature too. In isolated cases cataract can be a part of multisystem disorders. About 50 loci have been associated with hereditary cataract but genetics of many cataract remains unknown. Multi-gene panel and WES are helpful in studying mutations in disease-causing genes such as GEMIN4 that was found independently mutated with a syndrome of cataract, global developmental delay with/without renal involvement along with a syndrome that resembled galactosemia caused by mutations in CYP51A1. Researchers could identify a founder mutation in RIC1 (KIAA1432) in patients with cataract, brain atrophy, microcephaly with/without cleft lip and palate. For non-syndromic pediatric cataract, they could map a locus in a multiplex consanguineous family on 4p15.32 where sequencing revealed a truncating mutation in TAPT1 with biallelic inactivation of TAF1A and WDR87 indicating usefulness of clinical genomics[21]. Presenile cataract happens before 45 years of age while senile/age-related cataract happens thereafter. Congenital form is inherited in Mendelian fashion; however, age-related cataract tends to be multifactorial. DNA repair enzymes’polymorphisms can influence repair efficiency and may lead to cataract. Polymorphisms in AP endonuclease-1 (APE1)with APE1-141 T/T genotype and T allele frequencies were higher in cataract patients, while G/G genotype and G allele frequencies were lower in controls. APE1-141 G/G genotype has a protective role, and T allele has a deleterious role[22].About 1/3 of non-syndromic congenital cataracts are inherited.With an estimated prevalence of 1-6 cases per 10 000 live births, 50% congenital cataract cases have a genetic cause.Although congenital nuclear cataract can be caused by multiple factors, mutation remains most common cause. Interestingly,all three types of Mendelian inheritance have been reported in cataract; however, autosomal dominant transmission is common. Research on congenital cataract led to identification of several classes of genes that encode crystallins, lens-specific connexins, aquaporin, cytoskeletal structural proteins, and developmental regulators[23]. Major intrinsic protein of lens(MIP); known as AQP0, plays a role in transparency and development of lens. More than 10 mutations in MIP have been linked to cataracts but p.D150H turned be a novel diseasecausing mutation in MIP. Scientists have relied on NGS as an accurate, rapid, and cost-effective method in diagnosis of congenital nuclear cataract and identified a mutation,c.508dupC (p.L170fs) in MIP. This mutation resulted in a frame-shift as validated by Sanger[24-25]. Similarly, mutations like p.H277Y in connexin 50; Cx50 (GJA8) and p.P59L in gap junction protein alpha 3 genes (GJA3) were associated with congenital cataracts causing pulverulent nuclear cataracts in Chinese[26-37]. An oligomerization disrupting mutation,c.62G>A (p.R21Q), in crystallin alpha A (CRYAA) segregating in three generations of Australians (Caucasian) was detected.Haplotype analysis indicated a common ancestry between two South Australian families with this mutation thereby strengthening genotype-phenotype correlations between this functional mutation in CRYAA and pediatric cataract[38]. An insertion variant of CRYGD with nuclear cataract has also been identified (c.451_452insGACT) causing an autosomal dominant congenital cataract. Mutant protein is said to be cause of cataract with loss of solubility and localization to nucleus inside cells. Likewise, a heterozygous variant,c.3673G > A (p.V1225M) in periaxin (PRX) was identified in patients and two asymptomatic individuals of family in BGIShenzhen study[39-40]. Mutations in CRYAB gene (p.R11C and p.R12C) responsible for cataracts in consanguineous families have also been described. A perfect co-segregation of a mutation c.475C>T (p.Q155*) in exon 6 of CRYBB2 has been reported in Mexicans. Mutation in CRYBB2 seemed to be associated with pulverulent cataract with some variability[41-42].Sometime, genetic confirmation becomes necessary to establish cause and effect relationship despite results from other tests such as diagnosing patients with hypomyelination with 4H leukodystrophy with polymicrogyria and cataracts because of mutations in POLR3B encoding RNA polymerase III subunit[43]. A dominant D109A mutation in CRYAB was associated with myofibrillar myopathy, cardiomyopathy and cataract in a polish family. Change in CRYAB influenced both structural and functional aspects due to decreased stability of oligomers leading to aggregate formation and it appears that mutated RQDE sequence of CRYAB could impair CRYAB chaperone-like activity and promote aggregation of lens crystallins[44]. When analyzed for presence of SNP rs25487 from XRCC1 and relationship between risk factors such as smoking, alcohol intake, hypertension, and diabetes,comparison of genotype distribution in XRCC1 between presenile cataract group and group without cataract, then an increased risk of developing pre-senile cataract for genotype Gln/Gln in recessive inheritance models was observed after adjusting for above mentioned factors[45]. A miRNA-binding SNP rs2278414: G>A in DNA of 3'-terminal untranslated regions of ZNF350 was found associated with cataract through an altered miRNA regulation of ZNF350 in Chinese suggested age related susceptibility[46]. A splice donor site mutation c.2825+1G>A in EPHA2 co-segregated in a family resulting into a truncated product responsible for congenital cataract suggesting that WES can serve as an efficient strategy to scan variants in causative genes for heterogeneous diseases[47].
Glaucoma Glaucoma causes optic nerve damage and is referred as ‘sneak thief of sight’ because it can lead to permanent vision loss. Risk factors include family history, high blood pressure, eye injury and steroid abuse. It is common in African Americans and people over 60y. If left undetected, it can lead to blindness. So, experts recommend regular eye examinations.Being largest cause of irreversible blindness it affects more than 65 million people globally. It comprises of a heterogeneous group of conditions that damage retinal ganglion cells (RGC)and optic nerve. It can occur at any age but is common in older people. Susceptibility factors contribute to glaucoma and these factors belong to two groups: 1) those affecting intraocular pressure and 2) those modulating RGC activity and viability.Glaucoma can also be inherited as a Mendelian autosomal dominant/recessive trait, or as a complex trait. Different genes have been discovered with multiple interacting loci. Thus,genomic approaches are essential to study biology as they can provide clues about pathogenesis. Being heterogeneous primary open angle glaucoma (POAG), angle closure glaucoma and exfoliation syndrome (XFS) glaucoma as well as a few others associated with developmental abnormalities.POAG remains leading cause of blindness and to understand POAG, it is necessary to identify genes and their variants.CALM2 may be involved in the RGC death and oxidative damage to cell communication system and it may be responsible for pathogenesis of POAG[48]. Researchers identified MPP7 as a novel gene for POAG with its dysregulation under mechanical stress conditions[49]. Genomic regions known for glaucoma include defective genes such as myocilin, optineurin and CYP1B1 responsible for Mendelian transmission. SNPs in other genes are also associated with glaucoma. POAG ensues without any symptoms other than appearance of slow vision loss. Angle-closure glaucoma, although rare, is considered an emergency and symptoms include pain, nausea and visual disturbances. LOX1 linked with exfoliation type of glaucoma is caused by abnormal accumulation of protein in drainage system and other structures of eye were recently identified that was subsequently followed by CAV1/2 for POAG[50-51].Mutations in MYOC, PITX2, FOXC1, PAX6, CYP1B1, and LTBP2 can cause early onset (either congenital or juvenile)glaucoma. While these genes account for about 20% of cases with onset before age 40 years, for those individuals known to have a mutation, impact on clinical care and genetic counseling can be significant. Thus, appropriate screening via genetic testing and timely treatment of mutation carriers can prevent, or limit loss of eye sight. Detecting mutations in these genes can also identify mode of inheritance. Mutations in CYP1B1 and LTBP2 cause recessive disease, while glaucoma caused by all other known genes is inherited as a dominant trait[52]. Because variable expressivity and age are features of early onset, pattern of disease inheritance may not be clear in many cases. Defining disease causing mutation(s) also defines disease inheritance pattern and facilitates mutation carrier detection, making timely treatment strategies possible. Studies suggest that some individuals with early onset caused by mutations in MYOC could be treated in future with agents that limit effects of endoplasmic reticulum stress[53]. Gene testing for MYOC mutations could help identify individuals who might be eligible for clinical trials to test novel therapies for myocilin related glaucoma. Since there is no permanent cure,once someone develops this condition, he/she generally will need treatment for rest of life. If diagnosed early, vision loss due to glaucoma can be slowed or even prevented.
Inherited Optic Neuropathies Hereditary optic neuropathies comprise a group of disorders and may be caused by mitochondrial defects. It could be inherited as an autosomal recessive/dominant, X-linked, or in a maternal familial pattern. They often present as a painless feature with bilateral decreased vision that proceed slowly. Like Leber’s hereditary optic neuropathy (LHON) hereditary optic neuropathy may have an acute presentation characterized by bilateral decreased acuity, central scotomas, decreased color vision, and optic atrophy. Combination of family’s history, clinical presentation,and age of onset may distinguish disorders, but accurate diagnosis may be facilitated by genetic testing. Pathology is limited to RGC of optic nerve and is mediated by reduced oxidative phosphorylation, fragmentation of mitochondrial network, and increased sensitivity to apoptosis[54-55]. Two most common conditions seen are LHON and dominant optic neuropathy (DOA). LHON is common disorder characterized by a rapid, painless loss of vision in one eye which is subsequently followed by contra lateral eye in young males. Majority of cases are due to one of 3-point mutations of mtDNA affecting complex I or ND genes (G11778A,G3460A and T14484C). Since it is maternally inherited thus all offspring are bound to inherit mutations. DOA is an autosomal hereditary optic neuropathy and is commonest of inherited neuropathies. Unlike LHON, it shows no gender preference and develops as a slow, progressive, painless,and bilateral symmetric visual loss, beginning insidiously in first two decades of life. Although heterogeneous, a major locus has been mapped to chromosome 3q28 with a mutation in optic atrophy type 1 (OPA1)[56-57]. OPA1 is a dominantly inherited neuropathy and mutations in this gene encoding a dynamin related mitochondrial protein underlie optic atrophy because of truncations in exons 8 to 28 along with amino acid changes found in GTPase domain (exons 8 to 15)[58]. Mutation spectrum in the OPA1 gene can lead to marked heterogeneity resulting into lower mitochondrial DNA content and thus providing a direct evidence for its pathogenetic role[59]. Current knowledge of genetics of optic neuropathy makes it possible to test for mutations allowing pre-symptomatic testing and risk assessment. Ongoing advances have revealed important mechanisms that may also suggest potential therapeutic targets. Despite this, clinical manifestation from a broad range of optic nerve pathologies is difficult to differentiate from other diseases and for this reason genetic testing becomes important to determine basis of disease causation. Inherited retinal degeneration (IRD) is a group of heterogeneous diseases of which retinitis pigmentosa (RP) and LCA are most common and severe type. A group of 18 genes are known to cause LCA but it is not generally possible to identify causative genes through clinical examination[60]. Yet, patients with LCA and mutations in RPE65 could potentially benefit from current RPE65 gene based approaches. To ensure selection of therapy, testing for IRD genes will be necessary as more gene therapies are developed. Currently, most cases of inherited neuropathies are due to mutations in mitochondrial DNA causing LHON or mutations in nuclear gene OPA1 causing autosomal dominant optic atrophy, also known as Kjer optic neuropathy. Disease transmission is very different for these 2 conditions: mitochondrial DNA mutations causing LHON are inherited maternally, while OPA1 mutations are inherited as a dominant trait from either mother or father. In addition, both autosomal dominant optic atrophy and LHON exhibit variable expressivity within and among families, thus making it difficult to identify carriers. In such scenario, genetic testing is advised as it can confirm diagnosis and help define inheritance pattern.Additionally, patients with LHON may benefit from avoidance of certain environmental exposures that may contribute to overall disease risk[61].
Marfan Syndrome MFS is characterized by musculoskeletal abnormalities, cardiovascular diseases and ocular abnormalities. There is no cure for MFS. Patients with MFS are myopic, astigmatic and have ectopia lentis (EL)[62]. It is caused by genetic defects of connective tissue that has an autosomal dominant transmission. Mutations in fibrillin-1(FBN1) have been identified in MFS and Marfan-like disorders on chromosome 15 in q21.1 locus that encodes protein fibrillin. Isolated EL is due to mutations of FBN1 c.1948 C>T(p.Arg650Cys)[63-66]. Increased TGF-β1 is a risk factor and is caused by FBN1 mutations which lead to defects in signaling.FBN1 SNPs rs2118181 and rs1059177 do not cause MFS but are linked with dilatative pathology of aortic aneurysms(DPAA). Thus, TGF-β1 and FBN1 SNPs rs2118181 and rs1059177 could serve as biomarkers for diagnosis of DPAA.Presence of a single rs2118181 minor allele (G) increased amount of TGF-β1 by roughly one ng/mL. Two copies of FBN1 rs1059177 minor allele (G) were required to have an additive effect on TGF-β1. Men have higher TGF-β1 than women. Strong correlation between TGF-β1 and FBN1 SNPs suggests that a single nucleotide substitution in FBN1 might reduce bioavailability or binding properties of FBN1 and influence TGF-β1 activation and concentration. By establishing relationship between TGF-β1 and FBN1 SNPs rs2118181 and rs1059177 researchers provided evidence that their combinations might be used as biomarkers to identify patients at risk for aortic aneurysm and dissection[67]. Recently two mutations in MFS were identified including a frameshift insertion, p.G2120fsX2160 and a nonsense mutation,p.Arg529X (rs137854476) in FBN1 which may predispose for thoracic aneurysms/dissections, together with defects in ocular and skeletal systems[68]. Mutation p.C499S in calcium-binding epidermal growth factor (cbEGF)-like domain 3 of FBN1 and mutation p.C908Y were identified in an inter-domain region of hybrid motif 2 linked to cbEGF-like domain 10 that helped conclude that FBN1 mutations involving cysteine substitutions are associated with MFS and EL with MFS features. Also,pathology seemed serious when mutations disrupted three disulfide bridges in cbEGF-like domains which likely cause typical MFS than if mutations occurred in hybrid motifs[69].To rule out MFS physicians should include a positive history and manifestations such as body habitus, sub-luxated lenses or cardiac abnormalities. Diagnosis of bilateral lens luxation must be followed by complete examination and analysis of FBN1 to discard MFS due to severe systemic complications.Often, condition causes an aggressive secondary glaucoma that may require surgery with lensectomy, vitrectomy and drainage device implantation to avoid optic atrophy[70]. NGS is quite efficient in identifying lesions at exomic level which can be validated via Sanger. New variants, including a nonsense SNP in FBN1 and one missense mutation in exon 15 of LRP1 may be related to phenotype[71]. SNPs in transforming growth factor beta receptor 2 (TGFBR2) are associated with MFS and sudden death with coronary disease. Cardiovascular remodeling and T cell activation of TGFBR2 suggest that TGFBR2 SNP(s) is also related to coronary lesion[72]. Marfan-associated disorders are associated in TGFBR2 with mutation (K291K) caused by 873 C>T substitution suggesting spectrum of Marfanrelated disorders world-wide[73]. SNPs in folic acid metabolism enzyme are responsible for elevation of Hcy, correlating with pathogenesis of aneurysms and dissection. Increased level of Hcy was recently correlated with retinal layer thinning leading to retinopathy[74]. When Marfan patients were studied for SNPs of methylenetetrahydrofolate reductase (MTHFR; c.665C>T and c.1286A>C), methionine synthase (MTR; c.2756A>G)and methionine synthase reductase (MTRR; c.66A>G) it was noticed that severe cardiovascular involvement in Marfan patients, and especially aortic dissection was associated with higher Hcy and prevalence of homozygous genotypes of folic acid metabolism enzymes than mild or no cardiovascular involvement. Results suggested that impaired folic acid metabolism has an important role in development and aorta remodeling[75]. In a study it was reported that members in a family had EL, myopia and glaucoma but did not exhibit typical cardiovascular features of MFS but did have heterozygous missense mutation c.2368T>A; p.Cys790Ser in exon 19 in FBN1 further confirming worldwide FBN1 mutations[76].
Myopia Here image of an object is focused in front of retina.This is also associated with sight complications. Unfortunately,mechanism underlying pathogenesis is poorly understood. It is a complex disorder that can lead to blindness due to increased risk of premature cataracts, retinal detachment, glaucoma and macular degeneration. Genetics has been implicated in pathogenesis and it can affect up to 60% of some population and its development is influenced by genes and environment[77].Severe or high grade myopia is associated with ocular morbidities in form of retinal detachment, macular/choroidal degeneration, premature cataract, and glaucoma. Evidence documents heritability of non-syndromic forms especially for high grade myopia which is referred as myopic spherical refractive power of 5 to 6 diopters or higher. Studies could identify high grade and moderate myopia loci successfully[78].Past observations were derived from linkage studies but results from GWAS and sequencing identified additional loci/variants spanning all chromosomes. Some were linked with etiology itself such as locus on chromosome 8p23. Hepatocyte growth factor (HGF) and its receptor (C-MET) were previously reported with myopia in Asians. Caucasian dataset supported an association between mild to moderate myopia, HGF SNP rs3735520 and HGF haplotypes rs2286194, rs3735520,rs17501108 and rs12536657, rs2286194, and moderate association of extreme high myopia with rs2286194. However,C-MET SNPs with myopia in an Asian study were not replicated in Caucasians[79]. To ascertain position of a disease susceptibility gene around D21S0083i with whole genome case control analysis with 27 158 microsatellite markers in high myopia Japanese, SNP (rs2839471) was suggested to be in frequent recombinant region within UMODL1. This region might play role for susceptibility in myopia and warrants studies to know mechanism(s) by which UMODL1 contributes to myopia[80]. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) play important roles in scleral remodeling and are differentially expressed in myopia. Association of refractive error and SNPs in MMPs and TIMPs in old order Amish (AMISH) and Ashkenazi Jewish(ASHK) families revealed connections of ocular refraction to polymorphisms near MMP-1 and in MMP-2 in AMISH but not among ASHK families. Results suggested that MMP-1 and MMP-2 are involved in refractive variation in AMISH. Based on findings it can be assumed that genetic/environmental heterogeneity can contribute to differences between ethnic groups[81]. A functional SNP at 3’UTR of PAX6 can influence risk for myopia since allele SNP rs662702 demonstrated that C allele had significantly lower expression than did T allele.SNP rs662702 affects microRNA-328 binding site explaining differential effect through reduction in PAX6 protein which increases risk for myopia[82]. Interestingly, a progressive increase of myopia in a person affected with classical familial homocystinuria was diagnosed based on ophthalmological examination showing bilateral subluxation of lens with inferior and nasal displacement. Biochemistry detected increased amino acid (homocysteinemia) consistent with homocystinuria.Study showed compound heterozygous T353N and D444N mutations of cystathionine beta-synthase (CBS), and a C667T homozygous mutation of methylenetetrahydrofolate reductase(MTHFR). This study showed classical homocystinuria in patient’s father and sister, although they did not present systemic/ocular features. Therefore, it is necessary to rule out homocystinuria in EL patients, even in absence of systemic symptoms[83]. Evidence from myopia mapping (MYP3 locus),animal models and observations of glycemic control in humans suggests that insulin-like growth factor (IGF)-1 plays role in eye growth. This study was conducted to determine whether IGF-1 SNPs are associated with myopia in Caucasian high-grade myopia pedigrees. In all, three SNPs rs10860860,rs2946834, and rs6214 were studied and rs6214 showed association with high grade and any myopia after correction for multiple testing. Study supported a genetic association between IGF-1 and high-grade myopia. Findings were in line with evidence in a model showing that IGF-1 promotes ocular growth and axial myopia. Thus, IGF-1 may be a myopia gene and demands investigation[84]. In another study,variants showed positive association with extreme myopia with polymorphism of rs1242379 in IGF-1 to be associated with high myopia in Chinese[85]. So far more than 20 loci have been identified. High grade myopia locus MYP3 was reported on chromosome 12q21-23 by linkage having three genes:UHRF1BP1L, PTPRR, and PPFIA2 which were supported by ocular expression. They were found to be novel candidates for myopic development within MYP3 locus[86]. Interestingly,some associations have been validated and replicated in populations from different geographies and ethnicities[87].Mutations in P4HA2 encoding prolyl 4-hydroxylase 2 were also identified that were associated with non-syndromic high myopic condition[88].
Polypoidal Choroidal Vasculopathies PCV is characterized by branching of vascular network in choroid that terminates in polypoidal dilations. It can lead to serosanguinous detachments of retinal pigment epithelium (RPE) and subretinal hemorrhagic disorders that follow sub-retinal fibrosis.It may manifest earlier than other forms of disorders such as AMD. In principle, PCV is phenotypically like AMD and thus has been suggested to be a variant of AMD however PCV has a different history and response to treatments[89]. There are ethnic differences in disease prevalence as it is prevalent in African Americans and Asians[90-91]. Neovascular AMD(nAMD) and PCV; also refereed as a subtype of ‘wet’ AMD constitutes 55% of wet AMD in Asians where risk factors for PCV are unknown. Exudative AMD and PCV share similar choroidal vasculature, but responses to treatments are different.During investigation for haplotype associations tagging SNPs in C3 with both nAMD and PCV, and for potential epistatic effects on C3 SNP, rs17030 was associated with PCV. Further,interaction between rs17030 and gender was identified in PCV cases. After stratification by gender, rs17030 G allele was found to confer an increased risk in males but not in females. Haplotype AG defined by major alleles of rs17030 and rs344555 was associated with PCV in males. In contrast to PCV, none of eight other SNPs were associated with nAMD indicating that an association of C3 rs17030 with PCV in males and that C3 may have an epistatic effect with gender in pathogenesis of PCV[92]. Likewise, association of SNPs in serpin peptidase inhibitor clade G member 1 (SERPING1)with neovascular AMD and PCV demonstrated association with AMD in Caucasians but not in Asians providing an evidence for an ethnic diversity in etiology of AMD[93].One study assessed association of pigment epitheliumderived factor (PEDF) gene with AMD and PCV. However,polymorphisms showed no direct association indicating need for PEDF genotyping[94]. Meta-analysis of association with PCV and difference between PCV and nAMD revealed 31 SNPs in 10 loci that contributed to PCV. Among them,ARMS2-HTRA1 showed allelic diversity between PCV and nAMD confirming variants can affect phenotypic expressions of PCV and nAMD[95]. Exome sequencing of Han Chinese cohort followed by replication in four independent cohorts identified a rare c.986A>G (p.Lys329Arg) variant in FGD6 as significantly associated with PCV. Intracellular localization of FGD6-Arg329 is distinct from that of FGD6-Lys329. In vitro,FGD6 could regulate proangiogenic activity, and oxidized phospholipids increased expression of FGD6. FGD6-Arg329 promoted abnormal vessels in retina than FGD6-Lys329.Collectively, data suggested that oxidized phospholipids and FGD6-Arg329 act synergistically to increase susceptibility to PCV[96]. When complete HTRA1 and its promoter were sequenced in a Hong Kong Chinese cohort, it was revealed that rs11200638, c.34delCinsTCCT, c.59C>T, rs1049331 and rs2293870 were significantly associated. Notably, rs2672598 was significantly associated with exudative AMD. Logistic regression indicated that rs2672598 remained significant after adjusting for rs11200638 in exudative AMD. Moreover,rs11200638-rs2672598 joint genotype AA-CC conferred higher risk to exudative AMD. Promoter with rs2672598 C-allele showed higher luciferase than wild type T-allele, independent of rs11200638 genotype. Coherently, vitreous humor HTRA1 with rs2672598 CC genotype was significantly higher than that with TT genotype. Furthermore, rs2672598 C-allele was predicted to alter transcription factor binding sites, but not rs11200638 A-allele. Results revealed that HTRA1 rs2672598 is significantly associated with exudative AMD than PCV in ARMS2/HTRA1 region, and were responsible for elevated HTRA1 transcriptional activity and HTRA1 protein[97].Similarly, researchers investigated association of ATP-binding cassette, subfamily G, member 1 (ABCG1) with PCV and nAMD in independent Chinese and Japanese studies and have identified a new haplotype-tagging SNP, rs225396, in ABCG1 to be associated with PCV and nAMD in Chinese and Japanese providing a new evidence to support ABCG1 as susceptibility gene for PCV and nAMD[98]. When Meta-analysis was performed to examine effects of rs10490924 and rs11200638 at ARMS2/HTRA1 locus in PCV then it was observed that there was a strong association of ARMS2/HTRA1 locus with exudative AMD and PCV, suggesting two disorders shared similar mechanisms. Effect sizes indicate existence of additional genetic and environmental factors affecting them to different degrees[99]. In another study association of PCV,difference between PCV and AMD, and genotype-phenotype correlation of PCV was undertaken. LOC387715 rs10490924 was associated with PCV and its clinical manifestations, and showed a discrepant distribution between PCV and AMD.Variants in HTRA1, CFH, and C2 were also associated with PCV[100]. To investigate associations of C2-CFB-RDBPSKIV2L region with nAMD and PCV allele and haplotype frequencies of SNPs in the C2-CFB-RDBP-SKIV2L region were probed and results suggested that SKIV2L is likely a causal gene for nAMD, conferring a significant protection independent of CFH and HTRA1. These data do not support a role of this region in PCV, suggesting different mechanisms might be operating between nAMD and PCV[101]. Associations of variants in high density lipoprotein (HDL) metabolism with nAMD and PCV showed that CETP is a susceptibility gene for nAMD and PCV and that ABCG1 is putative gene for PCV. CETP exerts a modifying effect on CFH in genetic risk suggesting a link of HDL metabolism with neovascular AMD and PCV[102]. Interestingly, five SNPs on chromosomes 5, 8,9, 12 and 22 showed associations for disease manifestations.A known AMD associated SNP, CFH rs1329428, was among these top SNPs, while remainders have not been implicated.Evidence supports that symptomatic patients with PCV can have complete regression without vision loss with treatments as per recent mapping of PCV loci[95-96,103]. Like AMD, causes of PCV remain unknown[89].
Retinitis Pigmentosa People with retinitis pigmentosa (RP)experience gradual decline in vision because rods and cones start dying. RP is characterized by night blindness, narrowing of visual field, pigmentary changes or even alterations of retina,eventually leading to vision loss[104-106]. Retinal dystrophies including RP make a group of heterogeneous diseases caused by mutations in genes coding for proteins of cones and rods.RP consist of neurodegenerative disorders which are related to limitations of visual performances. During RP dystrophy,patients first experience night blindness and/or visual field constriction (secondary to rod dysfunctions), followed by alterations of central vision due to cone damage. During atypical form of RP (rod-cone dystrophy), cone’s functionalities are disrupted in comparison with rod’s ones. Diagnosis relies on documentation of loss in photoreceptor activity by electroretinogram and/or visual field testing. Prevalence of RP is variably reported in one case per 4000 individuals. Inherited dystrophies are epiphenomenon of a complex framework(syndromic RP), but often represent isolated disorder in 85%-90% of cases. Although more than 250 mutations have been detected in 100 of genes, defect is identifiable in 50% of cases only. Genotypes in RP are heterogeneous since a patient with same mutation may be affected by different phenotypes.RP can be inherited as autosomal dominant/recessive or as X-linked. Many forms are diagnosed in patients with no affected relatives. Dissecting clinico-genetic complexity has become an important objective of large scale projects[107].One approach is ‘deep sequencing’. It has identified several genes but a substantial unsolved case may have mutations that are known causes of retinal disease but not necessarily RP.Apparent discrepancy between defect and clinical findings warrants further evaluation[108]. In case of X-linked RP(XLRP), high mutation detection rate can make evaluation a valuable tool not only for counseling but also for prenatal testing. For example, it was discovered that proportion of RP2-mediated XLRP in Danish population is higher and proportion of RPGR-ORF15 is lower than reported thus strategies for diagnostics should consider population specific mutations. A phenotypic progression was reported for novel mutation in RPGR causing XLRP in people. Novel mutation in RPGR caused XLRP with complete penetrance. Affected females were highly myopic but retained better in visual functions than males. DNA analysis could identify a novel c.350G>A sequence in exon 5 of RPGR and it segregated with disease in families. Thus, data can be used to provide a mutation specific prognosis, and may also help recognize genotype[109].Clinically, RP1 gene mutations seem to be cause of autosomal dominant RP and mutations in RPGR remain frequent cause of XLRP. A five generation Swiss family with dominantly inherited RP caused by T494M mutation in precursor mRNA processing factor 3 (PRPF3) was characterized to relate phenotype to underlying mutation. A mutation in PRPF3 is a rare one compared to genes causing autosomal dominant RP but mutations in PRPF3 can cause a variable phenotype,unlike in previously described Danish, English, and Japanese families. This report was based on largest pedigree and provided a better understanding of phenotype-genotype description as caused by PRPF3 mutation[110]. More frequently involved gene in autosomal recessive RP code for subunits α and β of cGMP phosphodiesterase, RHO and cGMP gated ion channel CNGC. Similarly, mutations in other genes have been observed to cause RP and other retinal dystrophies[111].Whole exome NGS revealed missense mutation in hexokinase 1, HK1 c.2539G>A, p.Glu847Lys, tracking disease in all family members. One severely affected male was homozygous for this by linkage and had two copies of mutation. No other mutations were detected in linkage region nor were identified elsewhere in genome. Subsequent testing detected same mutation in four additional, unrelated families, for a total of five mutations in 404 probands. Out of 5 families, 3 were Acadian from Louisiana, 1 was French Canadian, and last one was Sicilian. Haplotype analysis chromosome in each family and homozygous individual revealed a rare, shared haplotype of 450 kb, suggesting an ancient founder mutation. HK1 is widely expressed, with multiple, abundant retinal transcripts,coding for H1K. The Glu847Lys mutation lies in a highly conserved site, outside of functional sites[112]. It is estimated that for retinal degenerations 40%-50% families have genetic basis for their diseases. When WGS was performed the evaluation revealed typical adolescent onset recessive RP.WGS identified 4 million variants in everyone. Two rare and deleterious compound heterozygous variants p. Arg281Cys and p.ARg487* were identified in ATP/GTP binding protein like 5 (AGBL5) as likely causal variants. Analysis confirmed segregation of variants with inherited dystrophies in pedigree.Homology models indicated destabilization of AGBL5 due to p.Arg281Cys change. Findings established involvement of mutations in AGBL5 in RP and validated WGS variant filtering pipeline analysis[113]. RP is endowed with highly varied consequences and progress in finding treatments is dependent on determining genes/mutations causing disease,which includes both gene discovery and mutation screening in patients and families. Despite complexities, current technology can detect mutations in 30%-80% of cases but finding treatments remain a challenge.
Stargardt’s Disease SD is a common form of juvenile macular dystrophy and is responsible for central vision loss in adults who are younger than 50y. Features of SD include progressive central visual loss, irregular yellow/white fundus flecks and atrophic macular lesions. Most inherit SD as an autosomal recessive disease, but it has also been reported as dominant trait. SD is caused by mutations in ABCA4 gene located on chromosome 1. More than 800 mutations in ABCA4 have been discovered causing autosomal recessive SD. Due to extensive heterogeneity variant associated phenotypes can manifest variability. Also, a high carrier frequency of pathogenic ABCA4 alleles in general population (about 1:20)results in pseudo-dominant inheritance thereby complicating diagnosis. When sequencing of ABCA4 was done in an unusual family for genotype/phenotype analysis with multiple macular phenotypes spanning across two generations with segregating four distinct ABCA4 alleles it found two known missense mutations; p.C54Y and p.G1961E. Comparative genomic hybridization (CGH) revealed a large deletion combined with a small insertion, c.6148-698_c.6670del/ins TGTGCACCTCCCTAG, and a new deep intronic variant,c.302+68C>T. Patients with p.G1961E mutation had mildest confined maculopathy with peripheral flecks while those with other alleles exhibited advanced state of retinal and choriocapillaries atrophy. This study epitomized genetic complexity of ABCA4 associated diseases as it contained variants from all classes of mutations in coding region, deep intronic, both single nucleotide and copy number variants accounting for phenotypes that tend to segregate in dominant fashion[114].Autosomal recessive macular dystrophy of childhood is caused by mutation(s) in retina specific ATP binding transporter gene (ABCR). Previously, ABCR cDNA and part of exonintron structure was described and discovery of a splicing mutation (571:2A>G) and missense mutation(s) in newly identified exons (R18W, R212C) provided additional support to broad heterogeneity. Genes involved in dominant form were mapped to chromosomes 13q (SD2) and 6q (SD3).One new kindred with dominant SD was identified in genetic linkage to SD3 locus. Because of a more severe macular phenotype in one patient in that family, gene responsible for recessive STGD1, ABCR was analyzed for variants in all family members. One allele of ABCR gene was shown to carry a stop codon generating mutation (R152X) in 3 members, including patient who had inherited dominant gene.A grandparent of that patient with same mutation developed AMD, consistent with observation that some variants in ABCR might increase susceptibility to AMD in heterozygous state. Based on this, it was proposed that there is a common pathway in macular degeneration that includes genes for both recessive and dominant form of SD. While describing genotype-phenotype correlation in one study with late onset,analysis of ABCA4 gene was performed using microarray analysis, sequencing, and multiplex ligation dependent probe amplification (MLPA). In addition, PRPH2 and CFH genes were also sequenced. As we know that late onset SD1 is at mild end of spectrum of retinal dystrophies caused by ABCA4 mutations because visual acuity is frequently preserved in late onset owing to foveal sparing. This phenotype may be caused by just 1 or 2 ABCA4 variants. One study aimed to identify genetics underlying severe retinal degeneration in one large family. Members were presented with early onset autosomal recessive RP and juvenile macular dystrophy.Records of family members were analyzed retrospectively, and ophthalmological and electrophysiological examinations were performed. Screening was done with microarrays followed by high density SNP genotyping, and segregation analysis. Two distinct phenotypes of dystrophy, LCA and SD were present in family; four patients with LCA were homozygous for a novel c.2557C>T (p.Q853X) mutation in CRB1, while two cases with SD, one was homozygous for c.5461-10T>C in ABCA4 and another was carrier of same mutation along with a novel ABCA4 mutation c.4773+3A>G. In a similar investigation when analysis of entire ABCA4 with SD was done, it revealed complex alleles with additional variants. Findings revealed that different mechanism(s) can lead to variable phenotypes within same family[115]. Yet, in another study a total of 36 alleles were identified. Two alleles were present in twelve out of 21 SD families, whereas in 4 out of 21 families only one allele was found. This work reported presence of 22 alterations,including two changes not found in other populations, c.2T>C(p.Met1Thr) and c.4036_4037delAC (p.Thr1346fs), and two novel disease associated variants, c.400C>T (p.Gln134X)and c.4720G>T (p.Glu1574X). Most of mutation(s) were missense. Seven frameshift variants, three nonsense mutations,and one splicing sequence changes were also found in SD chromosomes. However, most prevalent pathologic variant was missense mutation p.Leu11Pro representing high prevalence in comparison to other populations. Additionally, 23 SNPs were also identified including 4 intronic variants[116]. Differential diagnosis between late onset SD1 and AMD is challenging,therefore, a thorough clinical coupled with genetic analyses can make a distinction, which is important for counseling[117].When patients in family of Chinese descent were analyzed for exome of two patients, a total of 50 709 variations shared by patients were subjected to several filtering steps against existing databases. Identified variations were further verified in family by PCR and Sanger. Compound heterozygous variants p.Y808X and p.G607R of ATP binding cassette, sub-family A(ABC1), member 4 (ABCA4) gene, which encodes ABCA4 protein, a member of ATP-binding cassette (ABC) transport superfamily were successfully identified as causative mutations for SD. Findings helped provide one more novel ABCA4 mutation in Chinese with SD[118].
Uveal Melanoma Uveal melanoma (UM) is characterized by an uncontrolled proliferation in a clonal fashion because of genetic and epigenetic alterations. Signaling is dysregulated in UM in following way: 1) retinoblastoma pathway, because of cyclin D1 overexpression; 2) p53 signaling, because of MDM2 over-expression; 3) P13K/AKT pathway and 4) mitogen activated protein kinase/ERK pathway that are disturbed because of PTEN and GNAQ/11 mutations. Further,chromosomal abnormalities are common and include 6p gain,associated with a good prognosis, as well as 1p loss, loss of 3,and 8q gain, which correlate with high mortality. Abnormalities are identified by fluorescence in situ hybridization (FISH),CGH, microsatellite analyses, MLPA, and SNPs. UM can be categorized by expression profiling such as class 1 or class 2,latter correlating with poor survival, as do BRCA1 associated protein-1 (BAP1) inactivating mutations. Testing of UM has enhanced prognostication, especially when results are interpreted with patient’s history, histological findings and clinical data. Identification of abnormal pathways, genes and proteins in UM opens way for target based therapeutics,improving prospects for conserving vision and prolonging life[119]. Association between UM and cutaneous melanoma(CM) has been suggested, therefore individuals with a personal/family history should be screened. UM patients die of metastasis interventions. It arises from melanocytes in uveal tract. It is a primary malignant tumor in adults and remains common intraocular tumor in Caucasians[120]. Unfortunately,half UM patients develop metastasis which can be observed many years after treatment of primary tumor. In majority cases liver remains location of first manifestation. Based on chromosome 3 status UM can be divided into major classes that differ in metastatic potential: 1) tumors with a high risk to metastasis usually show monosomy 3, whereas 2) tumors showing disomy 3 rarely metastasize. If patients wish to know risk, prognostic testing of primary tumor can be done using biopsy. Genes involved in development such as SF3B1, BAP1,GNAQ, GNA11 and EIF1AX have been identified. Profiling in addition to chromosomal 3 analyses will refine classification/sub-classification and will influence diagnostics/therapeutics.Mutations in suppressor BAP1 are associated with increased risk for different tumors. Close family members of patients may be offered testing for BAP1[121]. In rare situations (young age, bilateral or multifocal forms) association with CM and/or familial aggregations of melanomas are indications of susceptibility. Unfortunately, UM has strong propensity to metastasize and prognosis remains poor. Options such as histopathology, pathway analyses, and genetic testing of UM have been suggested for monitoring and prognostic purposes.Among the newest, genetic testing has received attention as a powerful prognostic tool[122]. Thus, role of CDKN2A/P16INK4A, P14ARF and CDK4 germline mutations for predisposition was successfully carried out. Also, contribution of BRCA1/2 germline mutation(s) and a personal/family history of breast and ovarian cancers were evaluated. Results indicated that P14ARF, CDKN2A/P16INK4A, and CDK4 are not responsible for majority of cases. They also suggested that one case in a family with history of breast cancer was not sufficient to justify BRCA1/2 testing when classical criteria for testing were not present[123]. It is true that reported frequencies of mutations in melanoma are low in UM condition. However,number of families studied was limited and most studies used low sensitivity techniques for screening. Identifying frequency of alterations in melanoma with UM having increased risk is important for counseling. When screening for CDKN2A,p14ARF, and exon 2 of CDK4 was carried out it was revealed that variant (IVS1-69 C>T) in exon 1b of p14ARF in a patient and his mother also had UM. This study supported low frequency of germline mutation of CM in patients with UM with histories suggestive of a high risk for hereditary cancer.Therefore, testing for CDKN2A might be reserved for patients with family history of two or more CM cases[124]. MLPA using P027 assay was performed to determine whether extraocular extension of UM is representative of intraocular tumor growth with respect to copy number of chromosomes 1p, 3, 6, and 8.Tumors were micro-dissected and analyzed separately.Findings from this study suggested that biopsy may not be representative of underlying UM with respect to chromosome 1p, 3, 6, and 8q abnormalities. This indicated that both intraocular and extraocular parts of tumor should be sampled for accurate prognostic testing[125]. Germline sequence alterations in BAP1 gene with possible predisposition to hereditary cancer can predispose to UM, meningioma, lung carcinoma and possibly other cancers[126]. Monosomy 3 is linked with an aggressive, rapidly progressive disease while disomy or partial change of 3 and prominent mononuclear inflammatory infiltrate is associated with better prognosis[127].Loss of chromosome 3 is strongly associated with metastasis and has been proposed as basis for prognostic testing.However, it is not yet known whether techniques that identify loss of heterozygosity for chromosome 3 can also predict metastasis more accurately than those that detect only monosomy 3. To understand better, 53 UMs were analyzed by 28 SNPs across chromosome 3. SNPs were compared with FISH and CGH for metastasis. Prognostic tests based on SNPs,which detect chromosomal homologues and their sub-regions appears to be superior to methods that only detect changes in number of chromosomes. These observations could have implications to detect alterations in cancers with CGH based tools. Heterogeneity of chromosomal abnormalities of chromosomes 1, 3, 6, and 8 is present in almost UMs presentations. It was agreed that single random tumor biopsy/sample may not be true representative of whole tumor and,therefore, may be insufficient for prognoses[128]. A few candidates such as GNA11, GNAQ and DDEF1 have been proposed but in practice little is known about genetics of this disease[129]. To determine relationship between monosomy 3 and incidence of metastasis after testing UM using fine needle aspiration biopsy (FNAB). FNAB can be performed intraoperatively immediately before plaque radiotherapy and specimen can be used for testing using DNA amplification and microsatellite assay. According to FNAB results, patients having a complete monosomy 3 usually have poor prognosis than those with partial monosomy 3 or disomy 3. Also, patients with partial monosomy 3 do not differ in prognosis than those with disomy 3[130]. In following study researchers investigated utility of testing DNA from routinely stained and processed smears. Genotyping was carried out using 14 microsatellite markers on chromosomes 3, 6 and 8. Mutational screening in GNA11 and GNAQ was carried out by RFLP and results were compared with direct sequencing of frozen samples. DNAs extracted from 200 tumors were sufficient for reproducible testing of allelic imbalances and for studying somatic mutations in GNA11 and GNAQ. In conclusions, feasibility of utilizing stained smears from UM for testing using DNA is of sufficient quality to carry out genotyping for markers on chromosome 3, 6 and 8, as well as screening for somatic mutations in GNA11 and GNAQ[131]. Monosomy 3, 1p loss, 6q loss, and 8q and those classified under Class II by expression analysis are predictive of poor prognosis[132]. Due to shared epidemiological risk factors between CM and UM, researchers selected 28 SNPs identified as risk variants in previous GWAS on CM or CM related host phenotypes such as pigmentation and eye color and tested them for association with UM risk profile. By logistic regression analysis of 272 UM cases along with controls using an additive model, they identified 5 variants associated with UM risk. Three significantly associated variants rs12913832; rs1129038 and rs916977 were correlated and mapped at 15q12 in region of HERC2/OCA2,region responsible for eye color in humans. Data provided first evidence that genetic factors associated with pigmentation are risk loci for UM susceptibility[133]. Traditionally, clinicopathological features of these tumors were used to provide a limited prediction of metastatic risk. However, early studies using karyotype analysis, FISH and comparative hybridization identified multiple chromosomal abnormalities associated with higher risk of fatal metastasis. This correlation between specific abnormalities and a patient’s risk for development of metastasis has recently been studied, and development of new prognostic tests has allowed clinicians to predict metastatic risk with increased accuracy. Such novel tests include expression profiling, and MLPA, which detect deletions and amplifications of DNA in tumors[134]. Absence of BAP1 expression is associated with metastatic progression and reduced survival. In this study, investigators examined nuclear BAP1 (nBAP1) protein expression in primary UMs (PUMs)that show both typical and atypical clinical courses according to their chromosome 3 status, and secondary hepatic metastatic UM (MUM), correlating results with histological, clinical and survival data. Nuclear BAP1 expression was absent in 51%PUM patients, correlating strongly with poor prognostic clinico-pathological and genetic parameters and reduced survival. Lack of nBAP1 expression importantly identified a subset of ‘atypical’ PUM patients with disomy of chromosome 3 but with unexpected metastatic relapse. nBAP1 expression was absent in 77% MUM and expression was concordant in all paired PUM and MUM patients[135]. So far, results of prognostic testing in UM do not influence available therapeutic strategies.
DISCUSSION
Heritable and inflammatory diseases of retina such as AMD and RP are leading causes of blindness affecting one-third of people over age 75. AMD alone affects over 30 million,DR and glaucoma over 40 million and 65 million worldwide respectively. Most have strong genetic component. Advances have provided insights into genetic and pathophysiological mechanisms. NGS has revolutionized screening multiple genes and helped define mutations in approximately 20% with early onset glaucoma and approximately 50% of patients with optic atrophy and IRD[136]. Patients with disease without mutation in a known gene are likely to have mutation(s) in a novel gene(s)not yet discovered. Other techniques will be needed to detect abnormalities including karyotyping and MLPA. In real life situation role of genetics in clinical practice is not preferred domain as ophthalmologists tend to focus because school curriculum and ophthalmology residency place emphasis on routine diagnostics and surgical skills than on genetics. It should be mentioned that ophthalmology has played important roles in advancing impact of genomic medicine. It was retinoblastoma; first human gene to be cloned for its etiology.Leber hereditary optic neuropathy was first mitochondrial disorder to be studied in detail, and finally it was X-linked redgreen color deficiency which was again first X-linked disorder to be investigated[137]. Recently genetics has made resounding impact and as a result today’s ophthalmologists have working knowledge of genetic basis of diseases, and their clinical manifestations. Science has also evolved rapidly to ensure that genetic analysis is more accessible than it was in past.We hope genetic testing can improve accuracy of diagnoses and prognoses, can reduce risk of disease occurrences/recurrences, and facilitate development and delivery of disease mechanism specific care to patients. In US millions over 40 years of age are visually impaired and number is expected to triple by 2020[138]. Thus, effective surveillance, diagnosis and treatment will become increasingly important as population ages. Identification of risk factor(s) which contributes to diseases is first step towards development of screening tests and therapies. NGS, WGS, WES, and GWAS have identified elements/alleles for many disorders affecting vision and thus paved way for understanding genotypic-phenotypic correlations. Many conditions have ophthalmic manifestations and at same time also have genetic etiology. As elaborated in Figure 3, patients with eye diseases should be encouraged by ophthalmologists to undertake genetic testing. This would help promote ophthalmic genetics as an independent specialty. New developments have ushered interests in gene therapy and stem cells because of gratifying initial results. Despite these, there is paucity of animal models for complex diseases, including AMD and DR; because these diseases are not caused by single gene but involve complex interactions between genetics and environment, epigenetics, or other modes of influence. As we make further inroads in patient centric fashion wherein repertoire of genes shall be discovered mainly through patient’s samples, followed by selection of genes for detailed investigation for functions and pathways to reveal disease mechanism(s) to develop newer diagnostics and therapeutics.Despite progress that has been made so far, genetic testing is not yet recommended for many ocular disorders, but this will change if we move to clinical trials or treatments that are dependent on patients’ genotype. Interestingly,future application of genomic advances such as Clustered,Regularly Interspaced, Short Palindromic Repeats-associated Endonuclease 9 (CRISPR/Cas9) system sounds encouraging for ophthalmology. Progress in this could result in imminent applications since eye promises a favorable anatomical and immunological profile for genomic interventions and CRISPR/Cas9 can be integrated easily into ophthalmic care. CRISPR/Cas9 can precisely make editing in genome of cells including human embryos[139]. Other genome modification tools that were developed in past include Zinc-Finger Nucleases (ZFNs)[140]and transcription activator like effector nucleases (TALENs)[141]that can also enable permanent mutations by introducing double stranded breaks to activate repair pathways, but these are costly, time-consuming, and limit their widespread use.Thus, CRISPR/Cas9 appears simple for manipulating genomes.For example, Bassuk and colleagues tested whether CRISPR/Cas9 could be used in patient specific induced pluripotent stem cells (iPSCs) to precisely repair RPGR point mutation responsible for XLRP. Fibroblasts cultured from skin biopsy of XLRP patient were transduced to produce iPSCs carrying patient’s c.3070G > T mutation. iPSCs were then transduced with CRISPR guide RNAs, Cas9 endonuclease, and a donor homology template. Despite gene’s repetitive and GC-rich sequences, RPGR gene copies showed mutation correction and conversion to wild type allele. This was first successful report using CRISPR/Cas9 system to correct a pathogenic mutation in iPSCs from a patient with photoreceptor degeneration which clearly implied that cells can be successfully transformed into healthy retinal cells and then transplanted back into the same patient to treat vision loss[142]. This initial success with RP is an excellent example of how a new genome engineering technology could be applied for treating other forms of eye diseases that are caused by specific mutations that are directly responsible for vision loss. Despite these developments there remains an understandable concern too such as off-target mutagenesis/off-site genomic lesioning and perhaps mounting of an immune response against prolonged expression of Cas9 endonuclease in treated cells/organs of a patient. Thus, further improvements and careful screening for undesired mutations and development of a self-limiting CRISPR/Cas9 system could minimize duration of Cas9 expression. Such improvements will likely lead to clinical eye therapeutics of CRISPR/Cas9 system in future[143-144]. Interestingly, good thing about eye is that in comparison to other organs it is easy to monitor noninvasively, amenable for surgery and can accept modified cells as eyes are somewhat immune privileged. Nonetheless,there is still a long way to go but it is possible that one of first therapeutic uses of CRISPR/Cas9 system might be to develop it as a highly personalized approach for treating eye diseases both in humans and animals as demonstrated recently[145-146].
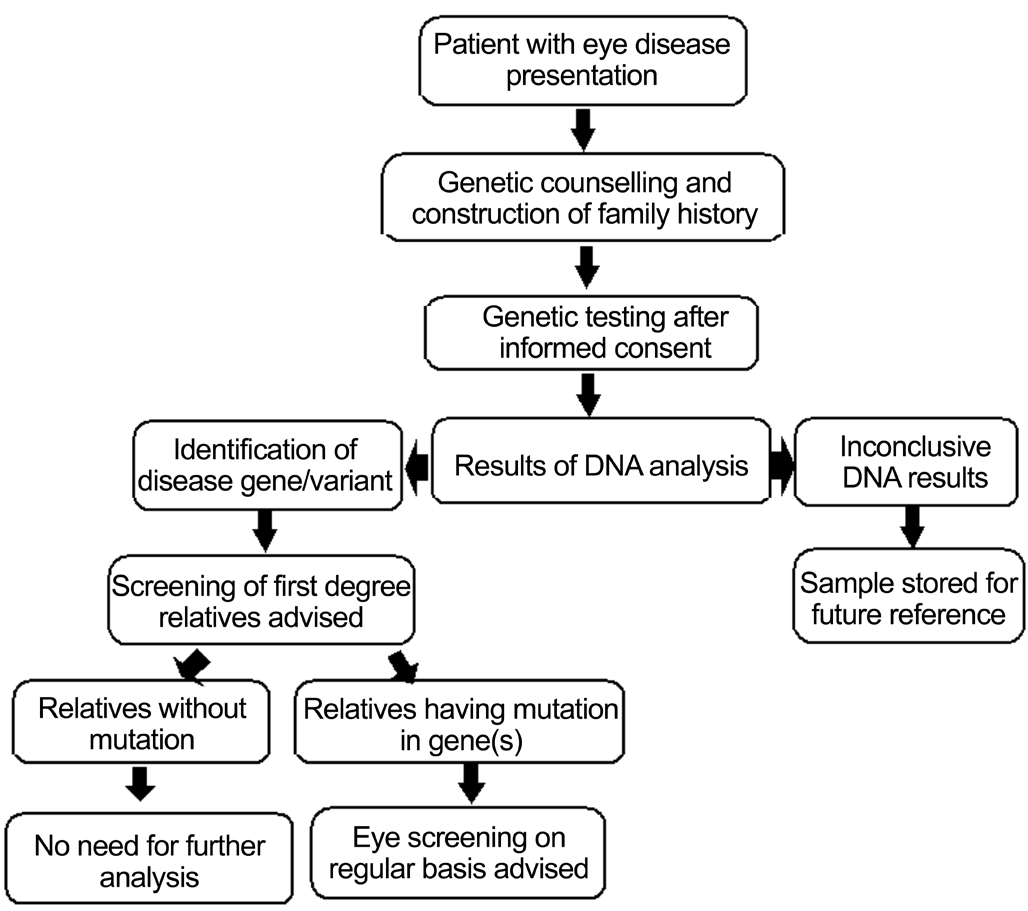
Figure 3 Flow chart for incorporating genomics care in eye diseases Management for genomic investigation, follow-up diagnostics and therapeutics become easier when an affected patient is subjected to a step-wise clinical care program.
CONCLUSION
Per American academy of ophthalmology task force recommendations genetic testing can truly make a positive mark on patients and their families. With so much happening and much more that is about to unfold, future does hold exciting possibilities. Driving these discoveries are rapid advances in infrastructure as well (e.g. the International HapMap Project for cataloging human genetic variations;http://www.hapmap.org), analytical methods, and medical biotechnology. This expansion in capabilities quickly has taken us from a genetics paradigm (where the influence of individual genes on health outcomes is paramount) to a genomics paradigm (where the complex influence of individual genes is considered, in concert, with each other and with the environmental exposures) on overall health outcomes.Here, we attempted to provide a concise but critical account of the ongoing genomic medicine studies exploring hereditary and inflammatory eye diseases linking risk variants/alleles that might offer avenues for developing suitable diagnostic modalities to test disease risk and therapeutic options.Identification of these and others to be discovered susceptibility loci can untangle the complex biological pathways underlying ocular pathophysiology pointing to new testable paradigms for treating and curing our patients. It is earnestly hoped that by utilizing these advanced investigative genomic approaches we may be able to provide affordable future clinical care for critical eye diseases.
ACKNOWLEDGEMENTS
We sincerely thank Aman Babbarwal and Karan Babbarwal for their excellent editing skills.
Foundations: Supported in part by NIH Heart, Lung, and Blood Institute (No.HLO74815); Institute of Neurological Disorders and Stroke (No.NS-084823).
Conflicts of Interest: Singh M, None; Tyagi SC, None.
REFERENCES
1 Shaw PX, Stiles T, Douglas C, Ho D, Fan W, Du H, Xiao X. Oxidative stress, innate immunity, and age-related macular degeneration. AIMS Mol Sci 2016;3(2):196-221.
2 Pang CP. Molecular genomics of eye diseases. Asia Pac J Ophthalmol(Phila) 2016;5(4):225-226.
3 den Hollander AI. Omics in ophthalmology: advances in genomics and precision medicine for Leber congenital amaurosis and age-related macular degeneration. Invest Ophthalmol Vis Sci 2016;57(3):1378-1387.
4 Sadagopan KA, Capasso J, Levin AV. Genetics for the ophthalmologist.Oman J Ophthalmol 2012;5(3):144-149.
5 Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY.Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis.Lancet Glob Health 2014;2(2):e106-e116.
6 Tan PL, Bowes Rickman C, Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics 2016;10(1):23.
7 Singh M, Olson P, Grossi F, Zhang Z, Tyagi N, Moshal KS, Tyagi SC.Differential expression of inflammatory cytokines and chemokines genes by homocysteine in the human retinal pigmented epithelial cells. FASEB J 2006;(20):A719-A719.
8 Priya RR, Chew EY, Swaroop A. Genetic studies of age-related macular degeneration: lessons, challenges, and opportunities for disease management. Ophthalmology 2012;119(12):2526-2536.
9 Ayala-Haedo JA, Gallins PJ, Whitehead PL, Schwartz SG, Kovach JL,Postel EA, Agarwal A, Wang G, Haines JL, Pericak-Vance MA, Scott WK. Analysis of single nucleotide polymorphisms in the NOS2A gene and interaction with smoking in age-related macular degeneration. Ann Hum Genet 2010;74(3):195-201.
10 Duvvari MR, Paun CC, Buitendijk GH, Saksens NT, Volokhina EB,Ristau T, Schoenmaker-Koller FE, van de Ven JP, Groenewoud JM,van den Heuvel LP, Hofman A, Fauser S, Uitterlinden AG, Klaver CC,Hoyng CB, de Jong EK, den Hollander AI. Analysis of rare variants in the C3 gene in patients with age-related macular degeneration. PLoS One 2014;9(4):e94165.
11 Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ,Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM,Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S,Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH)predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 2005;102(20):7227-7232.
12 Hoffman JD, Cooke Bailey JN, D'Aoust L, Cade W, Ayala-Haedo J, Fuzzell D, Laux R, Adams LD, Reinhart-Mercer L, Caywood L,Whitehead-Gay P, Agarwal A, Wang G, Scott WK, Pericak-Vance MA,Haines JL. Rare complement factor H variant associated with agerelated macular degeneration in the Amish. Invest Ophthalmol Vis Sci 2014;55(7):4455-4460.
13 Liu MM, Chan CC, Tuo J. Genetic mechanisms and age-related macular degeneration: common variants, rare variants, copy number variations,epigenetics, and mitochondrial genetics. Hum Genomics 2012;6:13.
14 Wang G, Dubovy SR, Kovach JL, Schwartz SG, Agarwal A, Scott WK, Haines JL, Pericak-Vance MA. Variants at chromosome 10q26 locus and the expression of HTRA1 in the retina. Exp Eye Res 2013;112:102-105.
15 Wang G, Spencer KL, Scott WK, Whitehead P, Court BL, Ayala-Haedo J, Mayo P, Schwartz SG, Kovach JL, Gallins P, Polk M, Agarwal A, Postel EA, Haines JL, Pericak-Vance MA. Analysis of the indel at the ARMS2 3'UTR in age-related macular degeneration. Hum Genet 2010;127(5):595-602.
16 Fritsche LG, Igl W, Bailey JN, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 2016;48(2):134-143.
17 Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J,Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308(5720):385-389.
18 Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005;308(5720):419-421.
19 Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science 2005;308(5720):421-424.
20 Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H,Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS,Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration.Science 2006;314(5801):992-993.
21 Patel N, Anand D, Monies D, Maddirevula S, Khan AO, Algoufi T,Alowain M, Faqeih E, Alshammari M, Qudair A, Alsharif H, Aljubran F, Alsaif HS, Ibrahim N, Abdulwahab FM, Hashem M, Alsedairy H,Aldahmesh MA, Lachke SA, Alkuraya FS. Novel phenotypes and loci identified through clinical genomics approaches to pediatric cataract. Hum Genet 2017;136(2):205-225.
22 Wang C, Lai Q, Zhang S, Hu J. Senile cataract and genetic polymorphisms of APE1, XRCC1 and OGG1. Int J Clin Exp Pathol 2015; 8(12):16036-16045.
23 Pichi F, Lembo A, Serafino M, Nucci P. Genetics of congenital cataract. Dev Ophthalmol 2016;57:1-14.
24 Shentu X, Miao Q, Tang X, Yin H, Zhao Y. Identification and functional analysis of a novel mip gene mutation associated with congenital cataract in a Chinese family. PLoS One 2015;10(5):e0126679.
25 Qin L, Guo L, Wang H, Li T, Lou G, Guo Q, Hou Q, Liu H, Liao S,Liu Z. A novel MIP mutation in familial congenital nuclear cataracts. Eur J Med Genet 2016;59(9):488-491.
26 Bennett TM, Mackay DS, Siegfried CJ, Shiels A. Mutation of the melastatin-related cation channel, TRPM3, underlies inherited cataract and glaucoma. PLoS One 2014;9(8):e104000.
27 Bennett TM, Maraini G, Jin C, Sun W, Hejtmancik JF, Shiels A.Noncoding variation of the gene for ferritin light chain in hereditary and age-related cataract. Mol Vis 2013;19:835-844.
28 Chacon-Camacho OF, Buentello-Volante B, Velazquez-Montoya R, Ayala-Ramirez R, Zenteno JC. Homozygosity mapping identifies a GALK1 mutation as the cause of autosomal recessive congenital cataracts in 4 adult siblings. Gene 2014;534(2):218-221.
29 Chograni M, Alkuraya FS, Maazoul F, Lariani I, Chaabouni-Bouhamed H. RGS6: a novel gene associated with congenital cataract, mental retardation, and microcephaly in a Tunisian family. Invest Ophthalmol Vis Sci 2014;56(2):1261-1266.
30 Ritchie MD, Verma SS, Hall MA, Goodloe RJ, Berg RL, Carrell DS,Carlson CS, Chen L, Crosslin DR, Denny JC, Jarvik G, Li R, Linneman JG, Pathak J, Peissig P, Rasmussen LV, Ramirez AH, Wang X, Wilke RA,Wolf WA, Torstenson ES, Turner SD, McCarty CA. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis 2014;20:1281-1295.
31 Thomas S, Thomas MG, Andrews C, Chan WM, Proudlock FA,McLean RJ, Pradeep A, Engle EC, Gottlob I. Autosomal-dominant nystagmus, foveal hypoplasia and presenile cataract associated with a novel PAX6 mutation. Eur J Hum Genet 2014;22(3):344-349.
32 Evers C, Paramasivam N, Hinderhofer K, Fischer C, Granzow M,Schmidt-Bacher A, Eils R, Steinbeisser H, Schlesner M, Moog U.SIPA1L3 identified by linkage analysis and whole-exome sequencing as a novel gene for autosomal recessive congenital cataract. Eur J Hum Genet 2015;23(12):1627-1633.
33 Mackay DS, Bennett TM, Culican SM, Shiels A. Exome sequencing identifies novel and recurrent mutations in GJA8 and CRYGD associated with inherited cataract. Hum Genomics 2014;8:19.
34 Niceta M, Stellacci E, Gripp KW, et al. Mutations Impairing GSK3-Mediated MAF Phosphorylation cause cataract, deafness, intellectual disability, seizures, and a Down Syndrome-like facies. Am J Hum Genet 2015;96(5):816-825.
35 Sun W, Xiao X, Li S, Guo X, Zhang Q. Exome sequencing of 18 Chinese families with congenital cataracts: a new sight of the NHS gene.PLoS One 2014;9(6):e100455.
36 Chen C, Sun Q, Gu M, Liu K, Sun Y, Xu X. A novel Cx50 (GJA8)p.H277Y mutation associated with autosomal dominant congenital cataract identified with targeted next-generation sequencing. Graefes Arch Clin Exp Ophthalmol 2015;253(6):915-924.
37 Wang L, Chen Y, Chen X, Sun X. Further evidence for P59L mutation in GJA3 associated with autosomal dominant congenital cataract. Indian J Ophthalmol 2016;64(7):508-512.
38 Javadiyan S, Craig JE, Souzeau E, Sharma S, Lower KM, Pater J,Casey T, Hodson T, Burdon KP. Recurrent mutation in the crystallin alpha A gene associated with inherited paediatric cataract. BMC Res Notes 2016;9:83.
39 Zhuang X, Wang L, Song Z, Xiao W. A novel insertion variant of crygd is associated with congenital nuclear cataract in a Chinese family.PLoS One 2015;10(7):e0131471.
40 Yuan L, Yi J, Lin Q, Xu H, Deng X, Xiong W, Xiao J, Jiang C, Yuan X,Chen Y, Deng H. Identification of a PRX variant in a Chinese family with congenital cataract by exome sequencing. QJM 2016;109(11):731-735.
41 Jiao X, Khan SY, Irum B, Khan AO, Wang Q, Kabir F, Khan AA,Husnain T, Akram J, Riazuddin S, Hejtmancik JF, Riazuddin SA.Missense mutations in CRYAB are liable for recessive congenital cataracts. PLoS One 2015;10(9):e0137973.
42 Messina-Baas O, Gonzalez-Garay ML, Gonzalez-Huerta LM, Toral-Lopez J, Cuevas-Covarrubias SA. Whole exome sequencing reveals a mutation in CRYBB2 in a large Mexican family with autosomal dominant pulverulent cataract. Mol Syndromol 2016;7(2):87-92.
43 Jurkiewicz E, Dunin-Wasowicz D, Gieruszczak-Bialek D, Malczyk K, Guerrero K, Gutierrez M, Tran L, Bernard G. Recessive mutations in POLR3B encoding RNA polymerase III subunit causing diffuse hypomyelination in patients with 4H leukodystrophy with polymicrogyria and cataracts. Clin Neuroradiol 2017;27(2):213-220.
44 Fichna JP, Potulska-Chromik A, Miszta P, Redowicz MJ, Kaminska AM, Zekanowski C, Filipek S. A novel dominant D109A CRYAB mutation in a family with myofibrillar myopathy affects alphaB-crystallin structure. BBA Clin 2017;7:1-7.
45 Lopez-Valverde G, Garcia-Martin E, Fernandez-Mateos J, Cruz-Gonzalez F, Larrosa-Poves JM, Polo-Llorens V, Pablo-Julvez LE,González-Sarmiento R. Study of association between pre-senile cataracts and rs11615 of ERCC1, rs13181 of ERCC2, and rs25487 of XRCC1 polymorphisms in a Spanish population. Ophthalmic Genet 2017;38(4):314-319.
46 Gu S, Rong H, Zhang G, Kang L, Yang M, Guan H. Functional SNP in 3'-UTR microRNA-binding site of ZNF350 confers risk for age-related cataract. Hum Mutat 2016;37(11):1223-1230.
47 Bu J, He S, Wang L, Li J, Liu J, Zhang X. A novel splice donor site mutation in EPHA2 caused congenital cataract in a Chinese family. Indian J Ophthalmol 2016;64(5):364-368.
48 Liu T, Xie L, Ye J, Liu Y, He X. Screening of candidate genes for primary open angle glaucoma. Mol Vis 2012;18:2119-2126.
49 Vishal M, Sharma A, Kaurani L, Alfano G, Mookherjee S, Narta K, Agrawal J, Bhattacharya I, Roychoudhury S, Ray J, Waseem NH,Bhattacharya SS, Basu A, Sen A, Ray K, Mukhopadhyay A. Genetic association and stress mediated down-regulation in trabecular meshwork implicates MPP7 as a novel candidate gene in primary open angle glaucoma. BMC Med Genomics 2016;9:15.
50 Wiggs JL, Kang JH, Yaspan BL, Mirel DB, Laurie C, Crenshaw A,Brodeur W, Gogarten S, Olson LM, Abdrabou W, DelBono E, Loomis S, Haines JL, Pasquale LR. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 2011;20(23):4707-4713.
51 Pasutto F, Krumbiegel M, Mardin CY, Paoli D, Lammer R, Weber BH, Kruse FE, Schlotzer-Schrehardt U, Reis A. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci 2008;49(4):1459-1463.
52 Fan BJ, Wiggs JL. Glaucoma: genes, phenotypes, and new directions for therapy. J Clin Invest 2010;120(9):3064-3072.
53 Zode GS, Bugge KE, Mohan K, Grozdanic SD, Peters JC, Koehn DR, Anderson MG, Kardon RH, Stone EM, Sheffield VC. Topical ocular sodium 4-phenylbutyrate rescues glaucoma in a myocilin mouse model of primary open-angle glaucoma. Invest Ophthalmol Vis Sci 2012;53(3):1557-1565.
54 Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet 2009;46(3):145-158.
55 Votruba M, Thiselton D, Bhattacharya SS. Optic disc morphology of patients with OPA1 autosomal dominant optic atrophy. Br J Ophthalmol 2003;87(1):48-53.
56 Cohn AC, Toomes C, Hewitt AW, Kearns LS, Inglehearn CF, Craig JE,Mackey DA. The natural history of OPA1-related autosomal dominant optic atrophy. Br J Ophthalmol 2008;92(10):1333-1336.
57 Toomes C, Marchbank NJ, Mackey DA, Craig JE, Newbury-Ecob RA, Bennett CP, Vize CJ, Desai SP, Black GC, Patel N, Teimory M,Markham AF, Inglehearn CF, Churchill AJ. Spectrum, frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet 2001;10(13):1369-1378.
58 Delettre C, Griffoin JM, Kaplan J, Dollfus H, Lorenz B, Faivre L,Lenaers G, Belenguer P, Hamel CP. Mutation spectrum and splicing variants in the OPA1 gene. Hum Genet 2001;109(6):584-591.
59 Kim JY, Hwang JM, Ko HS, Seong MW, Park BJ, Park SS.Mitochondrial DNA content is decreased in autosomal dominant optic atrophy. Neurology 2005;64(6):966-972.
60 Estrada-Cuzcano A, Roepman R, Cremers FP, den Hollander AI, Mans DA. Non-syndromic retinal ciliopathies: translating gene discovery into therapy. Hum Mol Genet 2012;21(R1):R111-R124.
61 Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog Retin Eye Res 2011;30(2):81-114.
62 Nahum Y, Spierer A. Ocular features of Marfan syndrome: diagnosis and management. Isr Med Assoc J 2008;10(3):179-181.
63 Faivre L, Collod-Beroud G, Callewaert B, et al. Clinical and mutationtype analysis from an international series of 198 probands with a pathogenic FBN1 exons 24-32 mutation. Eur J Hum Genet 2009;17(4):491-501.
64 Faivre L, Collod-Beroud G, Callewaert B, et al. Pathogenic FBN1 mutations in 146 adults not meeting clinical diagnostic criteria for Marfan syndrome: further delineation of type 1 fibrillinopathies and focus on patients with an isolated major criterion. Am J Med Genet A 2009;149A(5):854-860.
65 Faivre L, Masurel-Paulet A, Collod-Beroud G, et al. Clinical and molecular study of 320 children with Marfan syndrome and related type I fibrillinopathies in a series of 1009 probands with pathogenic FBN1 mutations. Pediatrics 2009;123(1):391-398.
66 Zhang L, Lai YH, Capasso JE, Han S, Levin AV. Early onset ectopia lentis due to a FBN1 mutation with non-penetrance. Am J Med Genet A 2015;167(6):1365-1368.
67 Sepetiene R, Patamsyte V, Zukovas G, Jariene G, Stanioniene Z,Benetis R, Tamosiunas A, Lesauskaite V. Association between Fibrillin1 polymorphisms (rs2118181, rs10519177) and transforming growth factor beta1 concentration in human plasma. Mol Med 2015;21(1).
68 Ma M, Li Z, Wang DW, Wei X. Next-generation sequencing identifies novel mutations in the FBN1 gene for two Chinese families with Marfan syndrome. Mol Med Rep 2016;14(1):151-158.
69 Li D, Yu J, Gu F, Pang X, Ma X, Li R, Liu N, Ma X. The roles of two novel FBN1 gene mutations in the genotype-phenotype correlations of Marfan syndrome and ectopia lentis patients with marfanoid habitus.Genet Test 2008;12(2):325-330.
70 Milla E, Leszczynska A, Rey A, Navarro M, Larena C. Novel FBN1 mutation causes Marfan syndrome with bilateral ectopia lentis and refractory glaucoma. Eur J Ophthalmol 2012;22(4):667-669.
71 Li G, Yu J, Wang K, Wang B, Wang M, Zhang S, Qin S, Yu Z. Exome sequencing identified new mutations in a Marfan syndrome family. Diagn Pathol 2014;9:25.
72 Choi YM, Shim KS, Yoon KL, Han MY, Cha SH, Kim SK, Jung JH.Transforming growth factor beta receptor II polymorphisms are associated with Kawasaki disease. Korean J Pediatr 2012;55(1):18-23.
73 Chen J, Li B, Yang Y, Hu J, Zhao T, Gong Y, Tan Z. Mutations of the TGFBR2 gene in Chinese patients with Marfan-related syndrome. Clin Invest Med 2010;33(1):E14-E21.
74 Srivastav K, Saxena S, Mahdi AA, Shukla RK, Meyer CH, Akduman L,Khanna VK. Increased serum level of homocysteine correlates with retinal nerve fiber layer thinning in diabetic retinopathy. Mol Vis 2016;22:1352-1360.75 Benke K, Agg B, Matyas G, Szokolai V, Harsanyi G, Szilveszter B,Odler B, Polos M, Maurovich-Horvat P, Radovits T, Merkely B, Nagy ZB, Szabolcs Z. Gene polymorphisms as risk factors for predicting the cardiovascular manifestations in Marfan syndrome. Role of folic acid metabolism enzyme gene polymorphisms in Marfan syndrome. Thromb Haemost 2015;114(4):748-756.
76 Micheal S, Khan MI, Akhtar F, Weiss MM, Islam F, Ali M, Qamar R, Maugeri A, den Hollander AI. Identification of a novel FBN1 gene mutation in a large Pakistani family with Marfan syndrome. Mol Vis 2012;18:1918-1926.
77 Simpson CL, Hysi P, Bhattacharya SS, Hammond CJ, Webster A,Peckham CS, Sham PC, Rahi JS. The Roles of PAX6 and SOX2 in Myopia: lessons from the 1958 British Birth Cohort. Invest Ophthalmol Vis Sci 2007;48(10):4421-4425.
78 Young TL. Molecular genetics of human myopia: an update. Optom Vis Sci 2009;86(1):E8-E22.
79 Yanovitch T, Li YJ, Metlapally R, Abbott D, Viet KN, Young TL.Hepatocyte growth factor and myopia: genetic association analyses in a Caucasian population. Mol Vis 2009;15(108):1028-1035.
80 Nishizaki R, Ota M, Inoko H, Meguro A, Shiota T, Okada E, Mok J,Oka A, Ohno S, Mizuki N. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye (Lond) 2009;23(1):222-229.
81 Wojciechowski R, Bailey-Wilson JE, Stambolian D. Association of matrix metalloproteinase gene polymorphisms with refractive error in Amish and Ashkenazi families. Invest Ophthalmol Vis Sci 2010;51(10):4989-4995.
82 Liang CL, Hsi E, Chen KC, Pan YR, Wang YS, Juo SH. A functional polymorphism at 3'UTR of the PAX6 gene may confer risk for extreme myopia in the Chinese. Invest Ophthalmol Vis Sci 2011;52(6):3500-3505.
83 Martinez-Gutierrez JD, Mencia-Gutierrez E, Gracia-Garcia-Miguel T, Gutierrez-Diaz E, Lopez-Tizon E. Classical familial homocystinuria in an adult presenting as an isolated lens subluxation. Int Ophthalmol 2011;31(3):227-232.
84 Metlapally R, Ki CS, Li YJ, Tran-Viet KN, Abbott D, Malecaze F,Calvas P, Mackey DA, Rosenberg T, Paget S, Guggenheim JA, Young TL.Genetic association of insulin-like growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci 2010;51(9):4476-4479.
85 Zhuang W, Yang P, Li Z, Sheng X, Zhao J, Li S, Yang X, Xiang W,Rong W, Liu Y, Zhang F. Association of insulin-like growth factor-1 polymorphisms with high myopia in the Chinese population. Mol Vis 2012;18:634-644.
86 Hawthorne F, Feng S, Metlapally R, Li YJ, Tran-Viet KN, Guggenheim JA, Malecaze F, Calvas P, Rosenberg T, Mackey DA, Venturini C, Hysi PG, Hammond CJ, Young TL. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Invest Ophthalmol Vis Sci 2013;54(3):2076-2086.
87 Rong SS, Chen LJ, Pang CP. Myopia Genetics-The Asia-Pacific Perspective. Asia Pac J Ophthalmol (Phila) 2016;5(4):236-244.
88 Guo H, Tong P, Liu Y, Xia L, Wang T, Tian Q, Li Y, Hu Y, Zheng Y, Jin X, Li Y, Xiong W, Tang B, Feng Y, Li J, Pan Q, Hu Z, Xia K.Mutations of P4HA2 encoding prolyl 4-hydroxylase 2 are associated with nonsyndromic high myopia. Genet Med 2015;17(4):300-306.
89 Kuo JZ, Wong TY, Ong FS. Genetic risk, ethnic variations and pharmacogenetic biomarkers in age-related macular degeneration and polypoidal choroidal vasculopathy. Expert Rev Ophthalmol 2013;8(2):127-140.
90 Imamura Y, Engelbert M, Iida T, Freund KB, Yannuzzi LA. Polypoidal choroidal vasculopathy: a review. Surv Ophthalmol 2010;55(6):501-515.
91 Laude A, Cackett PD, Vithana EN, Yeo IY, Wong D, Koh AH, Wong TY, Aung T. Polypoidal choroidal vasculopathy and neovascular agerelated macular degeneration: same or different disease? Prog Retin Eye Res 2010;29(1):19-29.
92 Liu K, Lai TY, Chiang SW, Chan VC, Young AL, Tam PO, Pang CP, Chen LJ. Gender specific association of a complement component 3 polymorphism with polypoidal choroidal vasculopathy. Sci Rep 2014;4:7018.
93 Liu K, Lai TY, Ma L, Lai FH, Young AL, Brelen ME, Tam PO, Pang CP, Chen LJ. Ethnic differences in the association of SERPING1 with age-related macular degeneration and polypoidal choroidal vasculopathy.Sci Rep 2015;5:9424.
94 Ma L, Tang SM, Rong SS, Chen H, Young AL, Kumaramanickavel G, Pang CP, Chen LJ. Association of PEDF polymorphisms with agerelated macular degeneration and polypoidal choroidal vasculopathy: a systematic review and meta-analysis. Sci Rep 2015;5:9497.
95 Ma L, Li Z, Liu K, Rong SS, Brelen ME, Young AL, Kumaramanickavel G, Pang CP, Chen H, Chen LJ. Association of genetic variants with polypoidal choroidal vasculopathy: a systematic review and updated Meta-analysis. Ophthalmology 2015;122(9):1854-1865.
96 Huang L, Zhang H, Cheng CY, et al. A missense variant in FGD6 confers increased risk of polypoidal choroidal vasculopathy. Nat Genet 2016;48(6):640-647.
97 Ng TK, Liang XY, Lai TY, Ma L, Tam PO, Wang JX, Chen LJ, Chen H, Pang CP. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep 2016;6:28639.
98 Sundaramurthy S, Swaminathan M, Sen P, Arokiasamy T, Deshpande S, John N, Gadkari RA, Mannan AU, Soumittra N. Homozygosity mapping guided next generation sequencing to identify the causative genetic variation in inherited retinal degenerative diseases. J Hum Genet 2016;61(11):951-958.
99 Liang XY, Lai TY, Liu DT, Fan AH, Chen LJ, Tam PO, Chiang SW,Ng TK, Lam DS, Pang CP. Differentiation of exudative age-related macular degeneration and polypoidal choroidal vasculopathy in the ARMS2/HTRA1 locus. Invest Ophthalmol Vis Sci 2012;53(6):3175-3182.
100 Chen H, Liu K, Chen LJ, Hou P, Chen W, Pang CP. Genetic associations in polypoidal choroidal vasculopathy: a systematic review and meta-analysis. Mol Vis 2012;18:816-829.
101 Liu K, Chen LJ, Tam PO, Shi Y, Lai TY, Liu DT, Chiang SW, Yang M,Yang Z, Pang CP. Associations of the C2-CFB-RDBP-SKIV2L locus with age-related macular degeneration and polypoidal choroidal vasculopathy.Ophthalmology 2013;120(4):837-843.
102 Liu K, Chen LJ, Lai TY, Tam PO, Ho M, Chiang SW, Liu DT, Young AL, Yang Z, Pang CP. Genes in the high-density lipoprotein metabolic pathway in age-related macular degeneration and polypoidal choroidal vasculopathy. Ophthalmology 2014;121(4):911-916.
103 Jin E, Bai Y, Huang L, Zhao M, Zhang C, Zhao M, Li X. Evidence of a novel gene HERPUD1 in polypoidal choroidal vasculopathy. Int J Clin Exp Pathol 2015;8(11):13928-13944.
104 Yuan Y, Zhou X, Wang F, Yan M, Ding F. Evidence for a novel autosomal dominant retinitis pigmentosa linked to chromosome 1p22.1-q12 in a Chinese family. Curr Eye Res 2011;36(2):154-167.
105 Garcia-Hoyos M, Garcia-Sandoval B, Cantalapiedra D, Riveiro R, Lorda-Sanchez I, Trujillo-Tiebas MJ, Rodriguez de Alba M, Millan JM, Baiget M, Ramos C, Ayuso C. Mutational screening of the RP2 and RPGR genes in Spanish families with X-linked retinitis pigmentosa.Invest Ophthalmol Vis Sci 2006;47(9):3777-3782.
106 Sullivan LS, Bowne SJ, Seaman CR, Blanton SH, Lewis RA,Heckenlively JR, Birch DG, Hughbanks-Wheaton D, Daiger SP. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 2006;47(10):4579-4588.
107 Chizzolini M, Galan A, Milan E, Sebastiani A, Costagliola C,Parmeggiani F. Good epidemiologic practice in retinitis pigmentosa: from phenotyping to biobanking. Curr Genomics 2011;12(4):260-266.
108 Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet 2013;84(2):132-141.
109 Al-Maskari A, O'Grady A, Pal B, McKibbin M. Phenotypic progression in X-linked retinitis pigmentosa secondary to a novel mutation in the RPGR gene. Eye (Lond) 2009;23(3):519-521.
110 Vaclavik V, Gaillard MC, Tiab L, Schorderet DF, Munier FL. Variable phenotypic expressivity in a Swiss family with autosomal dominant retinitis pigmentosa due to a T494M mutation in the PRPF3 gene. Mol Vis 2010;16:467-475.
111 Wang Y, Guo L, Cai SP, Dai M, Yang Q, Yu W, Yan N, Zhou X,Fu J, Guo X, Han P, Wang J, Liu X. Exome sequencing identifies compound heterozygous mutations in CYP4V2 in a pedigree with retinitis pigmentosa. PLoS One 2012;7(5):e33673.
112 Daiger SP, Sullivan LS, Bowne SJ, Koboldt DC, Blanton SH,Wheaton DK, Avery CE, Cadena ED, Koenekoop RK, Fulton RS, Wilson RK, Weinstock GM, Lewis RA, Birch DG. Identification of a novel gene on 10q22.1 causing autosomal dominant retinitis pigmentosa (adRP). Adv Exp Med Biol 2016;854:193-200.
113 Branham K, Matsui H, Biswas P, Guru AA, Hicks M, Suk JJ, Li H,Jakubosky D, Long T, Telenti A, Nariai N, Heckenlively JR, Frazer KA,Sieving PA, Ayyagari R. Establishing the involvement of the novel gene AGBL5 in retinitis pigmentosa by whole genome sequencing. Physiol Genomics 2016;48(12):922-927.
114 Lee W, Xie Y, Zernant J, Yuan B, Bearelly S, Tsang SH, Lupski JR,Allikmets R. Complex inheritance of ABCA4 disease: four mutations in a family with multiple macular phenotypes. Hum Genet 2016;135(1):9-19.
115 Jonsson F, Burstedt MS, Sandgren O, Norberg A, Golovleva I. Novel mutations in CRB1 and ABCA4 genes cause Leber congenital amaurosis and Stargardt disease in a Swedish family. Eur J Hum Genet 2013;21(11):1266-1271.
116 Maia-Lopes S, Aguirre-Lamban J, Castelo-Branco M, Riveiro-Alvarez R, Ayuso C, Silva ED. ABCA4 mutations in Portuguese Stargardt patients: identification of new mutations and their phenotypic analysis.Mol Vis 2009;15(59):584-591.
117 Westeneng-van Haaften SC, Boon CJ, Cremers FP, Hoefsloot LH,den Hollander AI, Hoyng CB. Clinical and genetic characteristics of lateonset Stargardt's disease. Ophthalmology 2012;119(6):1199-1210.
118 Zhou Y, Tao S, Chen H, Huang L, Zhu X, Li Y, Wang Z, Lin H, Hao F,Yang Z, Wang L, Zhu X. Exome sequencing analysis identifies compound heterozygous mutation in ABCA4 in a Chinese family with Stargardt disease. PLoS One 2014;9(3):e91962.
119 Coupland SE, Lake SL, Zeschnigk M, Damato BE. Molecular pathology of uveal melanoma. Eye (Lond) 2013;27(2):230-242.
120 Belmar-Lopez C, Mancheno-Corvo P, Saornil MA, Baril P, Vassaux G,Quintanilla M, Martin-Duque P. Uveal vs. cutaneous melanoma. Origins and causes of the differences. Clin Transl Oncol 2008;10(3):137-142.
121 Metz CH, Lohmann D, Zeschnigk M, Bornfeld N. Uveal melanoma:current insights into clinical relevance of genetic testing. Klin Monbl Augenheilkd 2013;230:686-691.
122 Werdich XQ, Jakobiec FA, Singh AD, Kim IK. A review of advanced genetic testing for clinical prognostication in uveal melanoma. Semin Ophthalmol 2013;28(5-6):361-371.
123 Buecher B, Gauthier-Villars M, Desjardins L, Lumbroso-Le Rouic L, Levy C, De Pauw A, Bombled J, Tirapo C, Houdayer C, Bressac-de Paillerets B, Stoppa-Lyonnet D. Contribution of CDKN2A/P16 (INK4A),P14 (ARF), CDK4 and BRCA1/2 germline mutations in individuals with suspected genetic predisposition to uveal melanoma. Fam Cancer 2010;9(4):663-667.
124 Abdel-Rahman MH, Pilarski R, Massengill JB, Christopher BN,Noss R, Davidorf FH. Melanoma candidate genes CDKN2A/p16/INK4A,p14ARF, and CDK4 sequencing in patients with uveal melanoma with relative high-risk for hereditary cancer predisposition. Melanoma Res 2011;21(3):175-179.
125 Lake SL, Damato BE, Dopierala J, Baudo MM, Taktak AF, Coupland SE. Multiplex ligation-dependent probe amplification analysis of uveal melanoma with extraocular extension demonstrates heterogeneity of gross chromosomal abnormalities. Invest Ophthalmol Vis Sci 2011;52(8):5559-5564.
126 Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB,Christopher BN, Boru G, Hovland P, Davidorf FH. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma,meningioma, and other cancers. J Med Genet 2011;48(12):856-859.
127 Abdel-Rahman MH, Cebulla CM, Verma V, Christopher BN, Carson WE 3rd, Olencki T, Davidorf FH. Monosomy 3 status of uveal melanoma metastases is associated with rapidly progressive tumors and short survival. Exp Eye Res 2012;100:26-31.
128 Dopierala J, Damato BE, Lake SL, Taktak AF, Coupland SE. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Invest Ophthalmol Vis Sci 2010;51(10):4898-4905.
129 Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC,Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R,Busam K, Speicher MR, O'Brien J, Bastian BC. Mutations in GNA11 in uveal melanoma. N Engl J Med 2010;363(23):2191-2199.
130 Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A,Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology 2011;118(2):396-401.
131 Christopher BN, Cebulla CM, Wakely PE Jr, Davidorf FH, Abdel-Rahman MH. Molecular genetic testing of uveal melanoma from routinely processed and stained cytology specimens. Exp Eye Res 2011;93(5):720-725.
132 Kaliki S, Shields CL, Shields JA. Uveal melanoma: estimating prognosis. Indian J Ophthalmol 2015;63(2):93-102.
133 Ferguson R, Vogelsang M, Ucisik-Akkaya E, Rai K, Pilarski R,Martinez CN, Rendleman J, Kazlow E, Nagdimov K, Osman I, Klein RJ,Davidorf FH, Cebulla CM, Abdel-Rahman MH, Kirchhoff T. Genetic markers of pigmentation are novel risk loci for uveal melanoma. Sci Rep 2016;6:31191.
134 Schopper VJ, Correa ZM. Clinical application of genetic testing for posterior uveal melanoma. Int J Retina Vitreous 2016;2(1):4.
135 Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer 2014;111(7):1373-1380.
136 Neveling K, Collin RW, Gilissen C, van Huet RA, Visser L, Kwint MP, Gijsen SJ, Zonneveld MN, Wieskamp N, de Ligt J, Siemiatkowska AM, Hoefsloot LH, Buckley MF, Kellner U, Branham KE, den Hollander AI, Hoischen A, Hoyng C, Klevering BJ, van den Born LI, Veltman JA,Cremers FP, Scheffer H. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat 2012;33(6):963-972.
137 Uthra S, Kumaramanickavel G. Gene therapy in ophthalmology.Oman J Ophthalmol 2009;2(3):108-110.
138 Cooke Bailey JN, Sobrin L, Pericak-Vance MA, Haines JL,Hammond CJ, Wiggs JL. Advances in the genomics of common eye diseases. Hum Mol Genet 2013;22(R1):R59-R65.
139 Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X,Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339(6121):819-823.
140 Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases.Nature 2005;435(7042):646-651.
141 Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T,Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 2011;39(21):9283-9293.
142 Bassuk AG, Zheng A, Li Y, Tsang SH, Mahajan VB. Precision medicine: genetic repair of retinitis pigmentosa in patient-derived stem cells. Sci Rep 2016;6:19969.
143 Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J.Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013;13(6):659-662.
144 Cabral T, DiCarlo JE, Justus S, Sengillo JD, Xu Y, Tsang SH.CRISPR applications in ophthalmologic genome surgery. Curr Opin Ophthalmol 2017;28(3):252-259.
145 Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol Ther 2017;25(2):331-341.
146 Sengillo JD, Justus S, Tsai YT, Cabral T, Tsang SH. Gene and cellbased therapies for inherited retinal disorders: an update. Am J Med Genet C Semin Med Genet 2016;172(4):349-366.