Table 1 Demographic data of studied participants
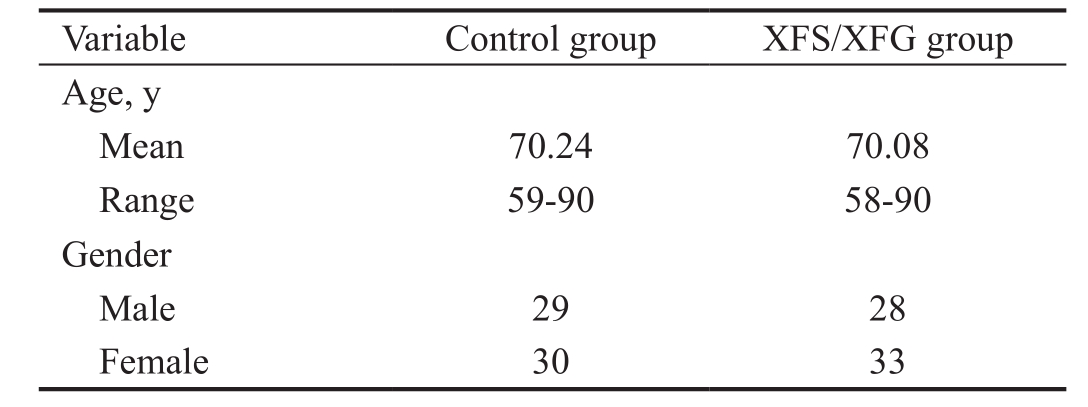
XFS/XFG: Exfoliation syndrome/glaucoma.
Wisam Shihadeh1,2, Omar Khabour3,4, Mohammed Bilal Khalil5, Alaa Al-Dabbagh1, Mustafa Al-Hashimi1
1Department of Ophthalmology/Special Surgery, Faculty of Medicine, Jordan University of Science & Technology, Irbid 22110, Jordan
2Department of Ophthalmology/Clinical Medical Sciences,Faculty of Medicine, Yarmouk University, Irbid 21163, Jordan
3Department of Medical Laboratory Sciences, Faculty of Applied Medical Sciences, Jordan University of Science &Technology, Irbid 22110, Jordan
4Faculty of Applied Medical Sciences, Taibah University, Al-Medina 42353, Saudi Arabia
5Department of Ophthalmology, Islami Hospital, Amman 11190, Jordan
Abstract
●AlM:To investigate the association between single nucleotide polymorphisms (SNPs) in theLOXL1gene with exfoliation syndrome/glaucoma (XFS/XFG) among Jordanians.
●METHODS:Sixty-one patients with XFS/XFG and 59 healthy control individuals were recruited in the study.Patients were diagnosed with XFS/XFG using standard clinical examination techniques. The exonic rs1048661 SNP and the intronic rs2165241 SNP inLOXL1gene were genotyped using sequencing technique. Allele and genotype frequencies were compared between cases and controls using Chi-square analysis.
●RESULTS:The G allele of the rs1048661 SNP and the T allele of the rs2165241 SNP were common in the sample with frequencies of 86.4% and 81.4%, respectively. ln addition, there were no significant differences in the genotypic and allelic distributions between patients and controls for rs1048661 SNP (P=0.770, OR=1.21, 95%Cl:0.56-2.60) and for rs2165241 SNP (P=0.605, OR=1.12,95%Cl: 0.59-2.09). ln addition, no significant associations were found between haplotypes of the examined SNPs and XFS/XFG in the sample (P>0.05).
●CONCLUSlON:Variations inLOXL1gene may not be associated with XFS/XFG in the Jordanian population.More studies are required to confirm the current findings.
●KEYWORDS:LOXL1; polymorphism; exfoliation syndrome;Jordan; glaucoma
Exfoliation syndrome (XFS) is a late onset fibrillopathic disease that involves formation and progressive accumulation of exfoliation material (XFM) in various ocular and extraocular tissues[1]. It is considered as the most global common cause of exfoliative glaucoma (XFG)[2-3]and it is associated with raised intraocular pressure (IOP) and increased aqueous humor outflow resistance[4]. On an ultrastructural level, XFM is a complex glycoprotein/proteoglycan structure composed of a protein core surrounded by abundant glycoconjugates[5-6]. The protein core comprises three components: 1) basement membrane component that contains laminin, nidogen, and fibronectin proteins; 2) components of the elastic fiber system that contains structures such as fibrillin-1 and elastin; 3) components with enzymatic activity such as matrix metallo-proteinases, clusterin chaperone, and the lysyl oxidase-like 1 (LOXL1)[7-8]. LOXL1 belongs to lysyl oxidase family of proteins that are involved in the biogenesis of connective tissue. LOXL1 has Cu-dependent amine oxidase activity that mediates the formation of the crosslinks in collagen and elastin[9]. In the ocular tissues, LOXL1 plays an essential role in homeostasis of elastic fiber networks[9-10].
In humans, LOXL1 is encoded byLOXL1gene that is located at 15q24.1[11]. In 2007, Thorleifssonet al[12]published the results of a genome-wide association study on Icelandic and Swedish populations and established a significant association between two exonic (rs3825942 and rs1048661) as well as one intronic (rs2165241) single nucleotide polymorphisms(SNPs) of theLOXL1gene with XFS/XFG. Both exonic SNPs,rs1048661 (R141L) and rs3825942 (G153D), are located in the chromosomal region 15q24.1, specifically in exon 1 ofLOXL1gene. This exon encodes the unique N-terminal domain that isrequired for proper LOXL1 enzyme activation and for substrate recognition and binding[13]. Several lines of evidence suggest that a reduction inLOXL1gene expression contributes to XFS/XFG development. In one study, using ocular tissue samples from the iris, lens and ciliary body,LOXL1expression was reduced by approximately 20% per risk allele of rs1048661[14].However, the association between theLOXL1SNPs and XFS/XFG has been examined in different populations worldwide,with contradictory results[15]. For example, rs1048661 G allele has been reported to increase the risk of XFG in Caucasian populations, whereas it has been shown to protect against the disease in Asian populations[16]. Therefore, in this study, we aimed to investigate the association ofLOXL1gene SNPs with XFS/XFG in Jordanian patients and to compare our data with the results from worldwide populations.
SubjectsThis cross-sectional analysis is part of an ongoing prospective study on XFS/XFG in Jordan. Sixty-one unrelated Jordanian patients with XFS/XFG and 59 age and gender matched unrelated controls from the same geographical area were included in the study. All patients were recruited from King Abdullah University Hospital, the major hospital that serves the northern municipalities of Jordan. XFS diagnosis was based on the presence of the characteristic XFM on the anterior lens surface on dilated ophthalmic examination by slit lamp biomicroscopy, regardless of IOP or glaucoma status.Exclusion criteria include patients presented with other related pathologies such as retinopathies, or maculopathies or other conditions causing secondary glaucoma[15,17]. Ethical approval on carrying out the study was obtained from the Institutional Review Board at Jordan University of Science and Technology.Informed consents were signed by all subjects enrolled in this study.
Demographics that include age, gender, time since diagnosis of the disease were obtained using a questionnaire. Informed consent was obtained from all XFS cases and normal subjects after the objectives and procedures of the study had been fully explained.
LOXL1Gene AnalysisA total of 5 mL of whole blood in EDTA-tube was collected from each participant and stored at-20.0℃. Genomic DNA was extracted from the blood samples using commercially DNA extraction kit (Promega, Madison,WI, USA) following the manufacturer’s instructions. For the rs2165241 SNP, DNA fragments were amplified using forward primer 5’-cgcattatagccatgcatca-3’ and reverse primer 5’-gtggccagaggtctgctaag-3’. For the rs1048661 SNP, DNA was amplified using forward primer 5’-gctcaactcgggctcaga-3’ and reverse primer 5’-ttctcgtactggctgacgaa-3’. Polymerase chain reaction (PCR) was carried out in a total of 25 µL volume containing ready-to-use master mix (Promega, Madison, WI,USA), 1 pmol of each primer, and 50 ng of genomic DNA. For both SNPs, the PCR conditions were 95℃ for 5min followed by 35 cycles of 95℃ for 60s, 60℃ for 35s and 72℃ for 45s,and the final extension at 72℃ for 10min. PCR products were then purified using quick-spin TM Kit (iNtRON Biotechnology Inc., Korea). The purified PCR products were sequenced using the Big Dye Terminator Cycle Sequencing Kit (version 3.1, Qiaquick, Germany). DNA sequencing was carried out in one direction in all exons using either the forward or the reverse primer. The DNA sequencing reaction was cleaned from the excess Dye Deoxy terminator using the DyeEx 2.0 spin purification kit (Qiagen, USA). The amplified DNA fragments were then sequenced using ABI 310 DNA sequencer(Applied Biosystems, Foster city, USA). Sequencing results were analyzed using chromasPro software (Technelysium,Australia). Reference sequences ofLOXLwere obtained from Ensembl Genome Browser (httb://www.ensembl.org/index.html).
Statistical AnalysisChi-square test, using SPSS software version 19 (SPSS Inc., Chicago, IL, USA), was carried out to evaluate genotype distribution and allele frequencies of the 2 studied polymorphisms for XFS cases and normal subjects.Haplotype frequencies were calculated using the SHEsis program (http://analysis.bio-x.cn/myAnalysis.php). AP-value of <0.05 was considered statistically significant.
A total of 61 patients with XFS/XFG and 59 healthy control subjects participated in the study. The average age in the exfoliation group was 70.08y (range 58-90) while the average age in the control group was 70.24y (range 59-90). Male to female ratio was 1:1.18 in the XFS/XFG group and 1:1.03 in the normal subject group (Table 1).
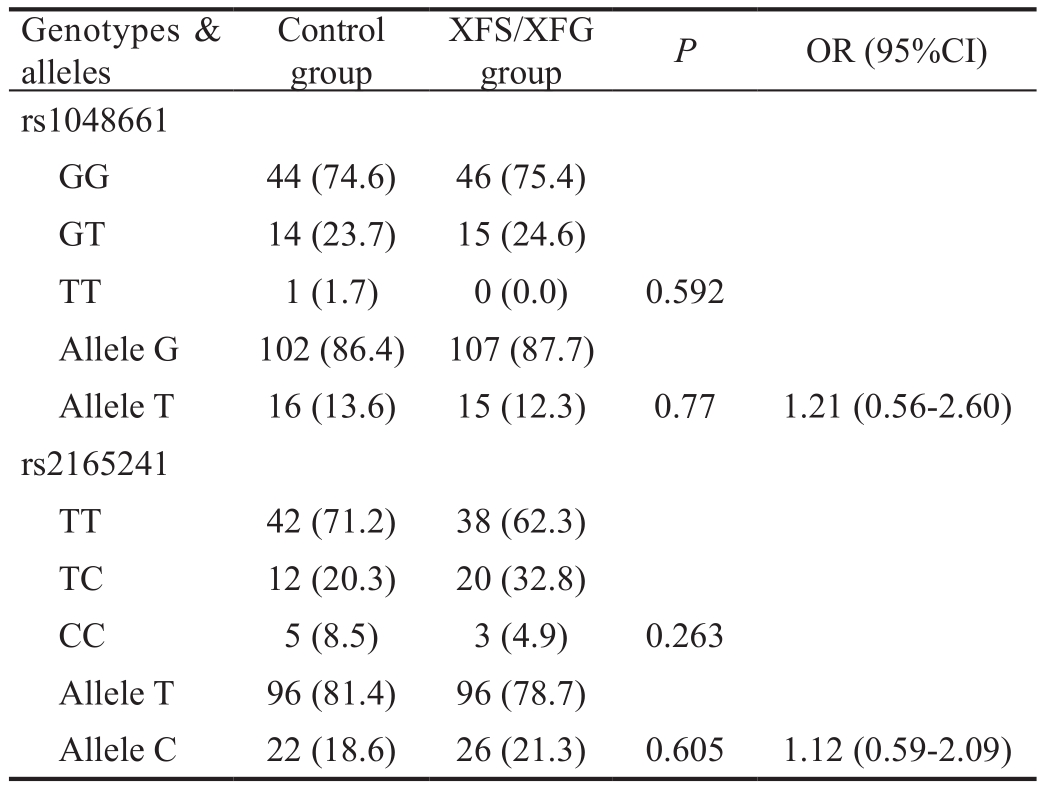
The G allele in the rs1048661 SNP and the T allele in the rs2165241 SNP were the most common alleles in the Jordanian population with frequencies of 86.4% and 81.4%, respectively.The abundance of these alleles is similar to that observed in other populations from other countries in the world (Table 2).Table 3 showed the allelic and genotypic distributions of the rs1048661 and rs2165241 SNPs of theLOXL1gene in both cases and controls. All examined SNPs are in Hardy-Weinberg equilibrium. There were no significant differences in the allelic distributions between patients and controls for rs1048661 SNP (P=0.770, OR=1.21, 95%CI: 0.56-2.60). In addition, no significant difference was obtained when genotypic distributionof the rs1048661 was considered (P=0.592). Similar findings were obtained with allelic distribution (P=0.605, OR=1.12,95%CI: 0.59-2.09) and genotypic distribution (P=0.263) of the rs2165241 SNP.
Table 1 Demographic data of studied participants

XFS/XFG: Exfoliation syndrome/glaucoma.
Table 2 Summary of the distribution ofLOXL1polymorphisms among various populations
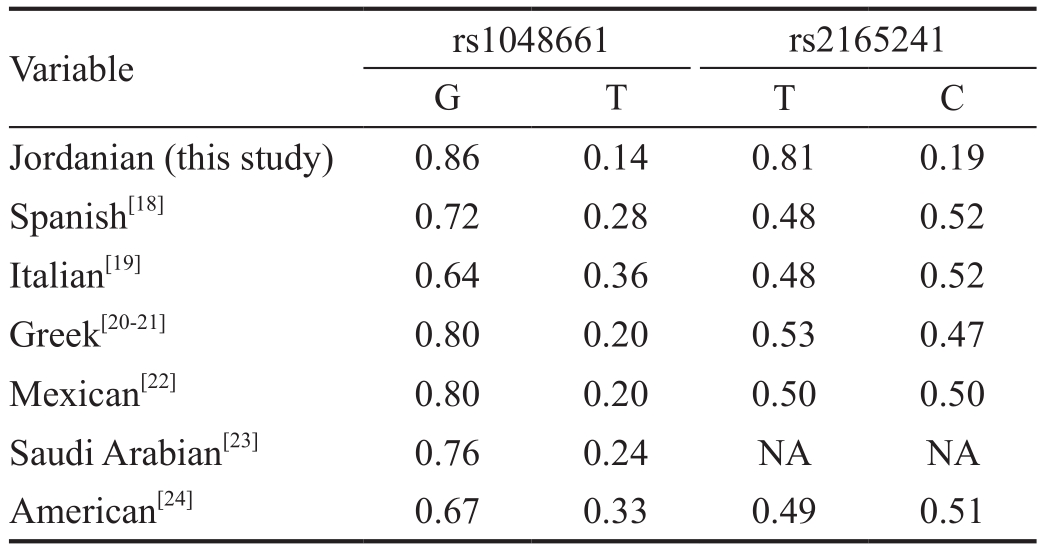
Table 3 Genotypic and allelic frequencies of rs1048661 and rs2165241 SNPs ofLOXL1in cases and controlsn(%)
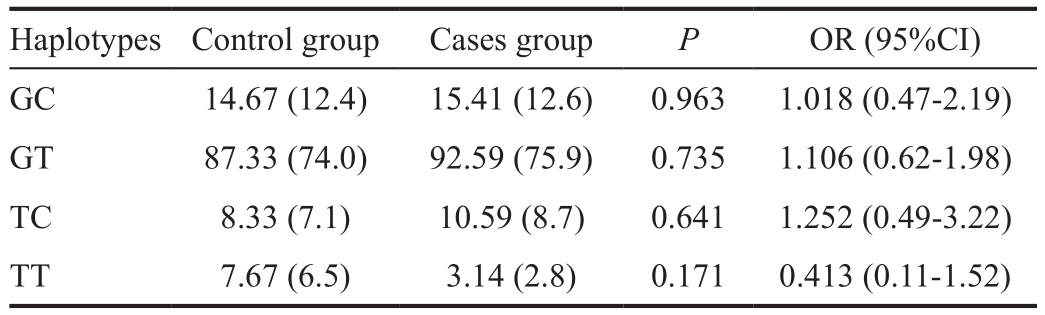
Haplotype analysis of the rs1048661 and rs2165241 SNPs was shown in Table 4. The most common haplotype in the population was GT (74%) followed by GC (12.4%), TC (7.1%)and TT (6.5%). The results showed no significant association between any of the examined haplotypes and the disease(GT:P=0.735, OR=1.106, 95%CI=0.62-1.98; GC:P=0.963,OR=1.018, 95%CI=0.47-2.19; TC:P=0.641, OR=1.252,95%CI=0.49-3.22; TT:P=0.171, OR=0.413, 95%CI=0.11-1.52). Thus, it appears that none of the examined SNPs are associated with XFS/XFG among Jordanians.
TheLOXL1gene belongs to the lysyl oxidase gene family which is involved in the biogenesis of connective tissue by formation of crosslinking in collagens and elastin[1]. Several studies have investigated the association between variations inLOXL1gene (particularly the rs1048661 and rs2165241 SNPs) and XFS/XFG, a condition where the extracellular spaces accumulate large amounts of cross-linked, amyloidlike fibrillar material and glycoproteins[15]. In this report, we compared distributions of genotypes, alleles and haplotypes ofrs1048661 and rs2165241 SNPs ofLOXL1genein XFS/XFG patients as well as normal subjects in Jordanians. The results showed absence of association between examined SNPs and the disease.
Table 4 Haplotype analyses of rs1048661 and rs2165241 SNPs ofLOXL1in cases and controlsn(%)
The rs1048661 is a nonsynonymous coding SNP located in the first exon of theLOXL1gene on chromosomal region 15q24.1[11]. The data showed no statistically significant difference between the XFS/XFG patients and control groups when comparing genotypic distributions and allelic frequencies of rs1048661. This finding is in agreement with previous reports conducted on Spanish[18], Italian[19], Greek[20-21],Mexican[22], Saudi[23], Amerian[24], Turkish[25-26]and Indian[27],Polish[28]and Chinese[29]populations. However, the G allele of rs1048661 SNP has been shown to be associated with XFS/XFG in Icelandic[12], Finnish[30]and Austrian[31]populations.
On the other hand, the G allele of rs1048661 SNP was found to be protective against the disease among Japanese[32-33]and Korean[34-35]populations. With respect to the intronic rs1048661 SNP, the results showed lack of association between rs2165241 SNP ofLOXL1geneand XFS/XFG among Jordanians. This result is in agreement with a previous report from Japan[32].However, the T allele was found to be associated with XFS/XFG in Caucasian populations such as Spanish[18], Polish[28],Icelandic[12], Finnish[30], German[19]and Austrian[31], while it was found to be protective against the disease among Chinese[36],Japanese[37]and Korean[34-35]populations. Thus, the relationship between rs1048661 and rs2165241 SNPs and XFS/XFG seems to be complex and might have a population specific component, being affected by the population specific gene pool as well as by gene-environment interactions. In support of this,allelic and genotypic distributions of the examined SNPs were found to be significantly different among different ethnicities.The allelic distribution of East Asian populations including Japanese and Chinese are reversed for rs1048661 and rs2165241 when compared with that of European populations(Table 2). In addition,in vitroassay has provided evidence that the rs1048661 SNP does not affect LOXL1enzymatic activity[38]. This means there are other regions in theLOXL1gene regulating the expression of the gene itself and/or that there are other genes or environmental factors contributing to the development of the disease[39]. Alternatively, combinations of SNPs (haplotypes) in theLOXL1gene might be more important in the determination of susceptibility to XFS/XFG rather than individual SNPs. In our study, none of the examined haplotypes were found associated with the disease.More studies at the molecular level are required to understand the role of different SNPs ofLOXL1gene in the pathogenesis of XFS/XFG.
In conclusion, we have performed an analysis of two SNPs inLOXL1gene and XFS/XFG in Jordanian population and have not found any association with the disease. More studies are required to confirm these findings.
Conflicts of Interest:Shihadeh W, None; Khabour O, None;Khalil MB, None; Al-Dabbagh A, None; Al-Hashimi M,None.
REFERENCES
1 Ritch R. Systemic associations of exfoliation syndrome.Asia Pac J Ophthalmol (Phila)2016;5(1):45-50.
2 Jiwani AZ, Pasquale LR. Exfoliation syndrome and solar exposure:new epidemiological insights into the pathophysiology of the disease.Int Ophthalmol Clin2015;55(4):13-22.
3 Pasquale LR, Borrás T, Fingert JH, Wiggs JL, Ritch R. Exfoliation syndrome: assembling the puzzle pieces.Acta Ophthalmol2016;94(6):e505-e512.
4 Ritch R. Ocular and systemic manifestations of exfoliation syndrome.J Glaucoma2014;23(8 Suppl 1):S1-S8.
5 Ritch R, Schlötzer-Schrehardt U, Konstas AG. Why is glaucoma associated with exfoliation syndrome?Prog Retin Eye Res2003;22(3):253-275.
6 Zenkel M, Schlotzer-Schrehardt U. Expression and regulation of LOXL1 and elastin-related genes in eyes with exfoliation syndrome.J Glaucoma2014;23(8 Suppl 1):S48-S50.
7 Ovodenko B, Rostagno A, Neubert TA, Shetty V, Thomas S, Yang A,Liebmann J, Ghiso J, Ritch R. Proteomic analysis of exfoliation deposits.Invest Ophthalmol Vis Sci2007;48(4):1447-1457.
8 Sharma S, Chataway T, Burdon KP, Jonavicius L, Klebe S, Hewitt AW,Mills RA, Craig JE. Identification of LOXL1 protein and apolipoprotein E as components of surgically isolated pseudoexfoliation material by direct mass spectrometry.Exp Eye Res2009;89(4):479-485.
9 Liu X, Zhao Y, Gao J, Pawlyk B, Starcher B, Spencer JA, Yanagisawa H,Zuo J, Li T. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein.Nat Genet2004;36(2):178-182.
10 Lucero HA, Kagan HM. Lysyl oxidase: an oxidative enzyme and effector of cell function.Cell Mol Life Sci2006;63(19-20):2304-2316.
11 Goy A, Gilles F, Remache Y, Zelenetz AD. Physical linkage of the lysyl oxidase-like (LOXL1) gene to the PML gene on human chromosome 15q22.Cytogenet Cell Genet2000;88(1-2):22-24.
12 Thorleifsson G, Magnusson KP, Sulem P,et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma.Science2007;317(5843):1397-1400.
13 Thomassin L, Werneck CC, Broekelmann TJ, Gleyzal C, Hornstra IK, Mecham RP, Sommer P. The pro-regions of lysyl oxidase and lysyl oxidase-like 1 are required for deposition onto elastic fibers.J Biol Chem2005;280(52):42848-42855.
14 Schlötzer-Schrehardt U. Molecular pathology of pseudoexfoliation syndrome/glaucoma-new insights from LOXL1 gene associations.Exp Eye Res2009;88(4):776-785.
15 Álvarez L, García M, González-Iglesias H, Escribano J, Rodríguez-Calvo PP, Fernández-Vega L, Coca-Prados M. LOXL1 gene variants and their association with pseudoexfoliation glaucoma (XFG) in Spanish patients.BMC Med Genet2015;16:72.
16 Aboobakar IF, Johnson WM, Stamer WD, Hauser MA, Allingham RR. Major review: exfoliation syndrome; advances in disease genetics,molecular biology, and epidemiology.Exp EyeRes2017;154:88-103.
17 Tang JZ, Wang XQ, Ma FF, Wang B, Wang PF, Peng ZX, Zhou XY. Association between polymorphisms in lysyl oxidase-like 1 and susceptibility to pseudoexfoliation syndrome and pseudoexfoliation glaucoma.PLoS One2014;9(3):e90331.
18 de Juan-Marcos L, Escudero-Domínguez FA, Hernández-Galilea E,Cabrillo-Estévez L, Cruz-González F, Cieza-Borrella C, Sánchez-Barba M, González-Sarmiento R. Association of lysyl oxidase-like 1 gene polymorphisms in pseudoexfoliation syndrome and pseudoexfoliation glaucoma in a Spanish population.Ophthalmic Genet2016;37(1):25-30.
19 Pasutto F, Krumbiegel M, Mardin CY, Paoli D, Lämmer R, Weber BH, Kruse FE, Schlötzer-Schrehardt U, Reis A. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma.Invest Ophthalmol Vis Sci2008;49(4):1459-1463.
20 Metaxaki I, Constantoulakis P, Papadimitropoulos M, Filiou E,Georgopoulos G, Chamchougia A, Papakonstantinou D, Markomichelakis N, Koutsandrea C, Halkiadakis I. Association of lysyl oxidase-like 1 gene common sequence variants in Greek patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma.Mol Vis2013;19:1446-1452.
21 Anastasopoulos E, Coleman AL, Wilson MR,et al. Association of LOXL1 polymorphisms with pseudoexfoliation, glaucoma, intraocular pressure, and systemic diseases in a Greek population. The Thessaloniki eye study.Invest OphthalmolVis Sci2014;55(7):4238-4243.
22 Jaimes M, Rivera-Parra D, Miranda-Duarte A, Valdés G, Zenteno JC.Prevalence of high-risk alleles in the LOXL1 gene and its association with pseudoexfoliation syndrome and exfoliation glaucoma in a Latin American population.Ophthalmic Genet2012;33(1):12-17.
23 Abu-Amero KK, Osman EA, Dewedar AS, Schmidt S, Allingham RR,Al-Obeidan SA. Analysis of LOXL1 polymorphisms in a Saudi Arabian population with pseudoexfoliation glaucoma.Mol Vis2010;16:2805-2810.
24 Challa P, Schmidt S, Liu Y, Qin X, Vann RR, Gonzalez P, Allingham RR, Hauser MA. Analysis of LOXL1 polymorphisms in a United States population with pseudoexfoliation glaucoma.Mol Vis2008;14:146-149.
25 Kasım B, İrkeç M, Alikaşifoğlu M, Orhan M, Mocan MC, Aktaş D.Association of LOXL1 gene polymorphisms with exfoliation syndrome/glaucoma and primary open angle glaucoma in a Turkish population.Mol Vis2013;19:114-120.
26 Tuncay FY, Aktas Z, Ergun MA, Ergun SG,et al. Association of polymorphisms in APOE and LOXL1 with pseudoexfoliation syndrome and pseudoexfoliation glaucoma in a Turkish population.Ophthalmic genet2017;38:95-97.
27 Ramprasad VL, George R, Soumittra N, Sharmila F, Vijaya L,Kumaramanickavel G. Association of non-synonymous single nucleotide polymorphisms in the LOXL1 gene with pseudoexfoliation syndrome in India.Mol Vis2008;14:318-322.
28 Malukiewicz G, Lesiewska-Junk H, Linkowska K, Mielnik M,Grzybowski T, Sulima N. Analysis of LOXL1 single nucleotide polymorphisms in Polish population with pseudoexfoliation syndrome.Acta Ophthalmol2011;89(1):e64-e66.
29 Lee KY, Ho SL, Thalamuthu A, Venkatraman A, Venkataraman D, Pek DC, Aung T, Vithana EN. Association of LOXL1 polymorphisms with pseudoexfoliation in the Chinese.Mol Vis2009;15:1120-1126.
30 Lemmelä S, Forsman E, Onkamo P, Nurmi H, Laivuori H, Kivelä T, Puska P, Heger M, Eriksson A, Forsius H, Järvelä I. Association of LOXL1 gene with Finnish exfoliation syndrome patients.J Hum Genet2009;54(5):289-297.
31 Mossböck G, Renner W, Faschinger C, Schmut O, Mossböck A,Weger M. Lysyl oxidase-like protein 1 (LOXL1) gene polymorphisms and exfoliation glaucoma in a Central European population.Mol Vis2008;14:857-861.
32 Fuse N, Miyazawa A, Nakazawa T, Mengkegale M, Otomo T, Nishida K. Evaluation of LOXL1 polymorphisms in eyes with exfoliation glaucoma in Japanese.Mol Vis2008;14:1338-1343.
33 Hayashi H, Gotoh N, Ueda Y, Nakanishi H, Yoshimura N. Lysyl oxidase-like 1 polymorphisms and exfoliation syndrome in the Japanese population.Am J Ophthalmol2008;145(3):582-585.
34 Park DY, Won HH, Cho HK, Kee C. Evaluation of lysyl oxidaselike 1 gene polymorphisms in pseudoexfoliation syndrome in a Korean population.Mol Vis2013;19:448-453.
35 Sagong M, Gu BY, Cha SC. Association of lysyl oxidase-like 1 gene polymorphisms with exfoliation syndrome in Koreans.Mol Vis2011;17:2808-2817.
36 Chen L, Jia L, Wang N, Tang G, Zhang C, Fan S, Liu W, Meng H,Zeng W, Liu N, Wang H, Jia H. Evaluation of LOXL1 polymorphisms in exfoliation syndrome in a Chinese population.Mol Vis2009;15:2349-2357.
37 Ozaki M, Lee KY, Vithana EN, Yong VH, Thalamuthu A, Mizoguchi T, Venkatraman A, Aung T. Association of LOXL1 gene polymorphisms with pseudoexfoliation in the Japanese.Invest Ophthalmol Vis Sci2008;49(9):3976-3980.
38 Kim S, Kim Y. Variations in LOXL1 associated with exfoliation glaucoma do not affect amine oxidase activity.Mol Vis2012;18:265-270.39 Wiggs JL, Pasquale LR. Expression and regulation of LOXL1 and elastin-related genes in eyes with exfoliation syndrome.J Glaucoma2014;23(8 Suppl 1):S62-S63.
Citation:Shihadeh W, Khabour O, Khalil MB, Al-Dabbagh A, Al-Hashimi M. Association ofLOXL1gene common sequence variants in Jordanian patients with exfoliation syndrome and exfoliative glaucoma.Int J Ophthalmol2018;11(10):1583-1587
DOl:10.18240/ijo.2018.10.02
Accepted:2018-08-09
Received:2017-08-11
Correspondence to:Wisam Shihadeh. Faculty of Medicine,Yarmouk University, PO Box 150474, Irbid 21141, Jordan.wshihadeh@yu.edu.jo