Effects of miR-22 on viability, migration, invasion and apoptosis in retinoblastoma Y79 cells by targeting highmobility group box 1
Min Liu1,2, Shuo-Min Wang3, Zheng-Xuan Jiang1, Hennein Lauren4, Li-Ming Tao1
1Department of Ophthalmology, the Second Affiliated Hospital of Anhui Medical University, Hefei 230601, Anhui Province,China
2Department of Ophthalmology, the Fourth Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui Province,China
3Department of Oncology, the Fourth Affiliated Hospital of Anhui Medical University, Hefei 230032, Anhui Province,China
4Department of Ophthalmology, University of California, San Francisco 94143-0730, California, USA
Abstract
●AlM:To explore the effect of miR-22 on viability, migration,invasion and apoptosis in retinoblastoma (RB) Y79 cells and to further detect the potential mechanism.
●METHODS:Plasmids were constructed to change the expression level of miR-22 in Y79 cells. Real-time reverse transcription polymerase chain reaction (RT-PCR) was conducted to test the expression level of miR-22. After changing the expression of miR-22, the mRNA and protein levels of high-mobility group box 1 (HMGB1) were investigated using RT-PCR and Western blotting. The effect of miR-22 on viability was analyzed by using cell counting kit-8 (CCK-8) assay and the effect on apoptosis was detected by the flow cytometry. Wound healing migration assay and Transwell invasion assay were used to detect the effects of miR-22 on cell motility.
●RESULTS:miR-22 inhibited viability, migration and invasion, while promoting apoptosis, in RB Y79 cells. The inhibition rate of miR-22 overexpression group at 12, 24,48h was 11.71%±2.54%, 21.36%±1.39% and 29.44%±1.15%,respectively. Cellular apoptosis was higher in miR-22 overexpression group (17.00%±0.39%) compared with negative control (4.38%±0.38%). miR-22 negatively mediated the expression of HMGB1. Furthermore, decreased HMGB1 significantly attenuated viability, migration and invasion, while promoting apoptosis. Enforced expression of HMGB1 partially rescued the effects of miR-22 overexpression on cell viability, migration, invasion and apoptosis. Moreover, the phosphorylated protein kinase B (p-AKT) was significantly downregulated in the HMGB1 shRNA group and miR-22 overexpression group and elevated in the HMGB1 overexpression group compared with the normal control.
●CONCLUSlON:miR-22 inhibites viability, migration and invasion and increases apoptosis in Y79 cells by targeting HMGB1. These findings may provide a therapeutic strategy for RB.
●KEYWORDS:miR-22; viability; migration; invasion;apoptosis; retinoblastoma; HMGB1
INTRODUCTION
Retinoblastoma (RB) is one of the most common intraocular malignant tumors in infants and young children. The incidence of RB is approximately one case in every 15 000 to 20 000 live births. Early detection and timely treatment of RB have led to a 95% survival rate in developed countries;however, in less developed countries the mortality rate is as high as 70%[1-3]. This may be in part due to delays in diagnosis, as most patients are diagnosed during the advanced stages of RB, especially in developing countries[4]. Despite improvements in RB treatment, the molecular mechanisms underlying its pathology are not fully understood. It is helpful to explore the molecular mechanisms of RB initiation in the design of new targeted therapy.
miRNAs, short non-coding RNAs that regulate up to one third of protein-coding gene expression by inducing mRNA degradation or translational repression[5-8]. miR-22, a 22 nucleotide non-coding RNA, is initially cloned in HeLa cells[9]and serves an essential role in a variety of cancers types[10-11].
Recently, miR-22 was found to dysregulate in many kinds of cancer, being downregulated in glioblastoma[12], gastric cancer[10]and hepatocellular carcinoma[13], and upregulated in prostate cancer[14]. Moreover, miR-22 expression is downregulated in human RB, also in mouse and human RB cell lines[15]. However, the function and molecular mechanism of miR-22 are yet to be elucidated in RB.
In addition, miR-22 has been reported to inhibit cell proliferation and migration through HMGB1 in osteosarcoma[11]. As an extracellular molecule, HMGB1 is implicated in cell differentiation, inflammation, cell migration and tumor progression by binding to advanced glycation end product receptors and Toll-like receptors[16-17]. HMGB1 is overexpressed in various kinds of cancers including gastric cancer[18], breast cancer and melanoma[19-20]. Recent studies have demonstrated that HMGB1 is correlated with optic nerve invasion and poor differentiation in RB[21]. These findings motivated the examination of the role of miR-22 in HMGB1 expression in RB.
In our present work, we investigated the role of miR-22 in viability, migration, invasion and apoptosis in RB Y79 cells and further detected the target gene HMGB1, with an attempt to provide novel treatment strategies to prevent the metastasis of RB.
MATERIALS AND METHODS
Cell CultureThe human RB cell line Y79 was obtained from Beina Chuanglian Biotechnology Institute (Beijing, China).After resuscitation, the cells were preserved in RPMI1640 medium (Gibco, USA) containing 10% fetal bovine serum(FBS) and 1% penicillin/streptomycin and were placed in a 37℃, 5% CO2incubator. We changed the medium every two to three days.
Construction of Plasmids and the Experiment GroupsThe following eukaryotic plasmids were all constructed by the Gene Pharma Company (Shanghai, China). The present study used three groups in the stable transfection experiment with miR-22 overexpression: the normal control, negative control(GV251) and miR-22 overexpression transfection vector group(GV251/miR-22). Three groups were also designated in the stable transfection experiment with miR-22 RNA interference recombinant plasmid: the normal control, negative control(GV249) and miR-22 RNA interference recombinant plasmid(GV249/miR-22-siRNA).
In addition, there were three groups in the stable transfection experiment with HMGB1 overexpression: the normal control,negative control (GV230) and HMGB1 overexpression transfection vector groups (GV230/HMGB1). Three groups were also designated in stable transfection experiment with HMGB1 RNA interference recombinant plasmid: the normal control, negative control (GV102) and HMGB1 RNA interference recombinant plasmid group (GV102/HMGB1-siRNA).
TransfectionWhen the cells confluence degree reached 90%,cells were plated at 1×106cells/well in 6-well plates with antibiotics free medium. Transfection of plasmids into the cells was carried out using Lipofectamine®2000 (Invitrogen, USA).Subsequently, cells were collected for further experiments.
Cell Viability AssayFollowing transfection, enhanced cell counting kit-8 assay (CCK-8; Beyotime, China, CK0041) was used to evaluate cell viability. Cells were seed in 96-well plates and then 10 μL of CCK-8 reagent was added to the samples for 2h. The absorbance value of each well was detected using a spectrophotometer (Thermo, USA) at 450 nm (A450). Three independent experiments were performed for each case.
Wound-healing Migration AssayThe cells were incubated in 6-well plates. After being grown to 90% confluence, cells were scratched using a 200 μL micropipette. Images were captured by a microscope in five predetermined fields (×100) at 0, 24 and 48h. The distance between the cells was then measured and quantified according to cell migration =(initial scratch width at 0h—final scratch width at 24 or 48h)/initial scratch width at 0h.
Transwell Invasion AssayInvasion chambers (Costar,USA) were utilized to evaluate cell invasion ability. The 2×103cells were plated at the upper chamber with serumfree medium. There are RPMI1640 medium with 10% FBS in the lower chambers, and the filters of the chambers were coated with Matrigal (BD, USA). After 24h, invaded cells were at the bottom of the chambers, they were fixed with 4%paraformaldehyde and then stained with hematoxylin. Images were acquired by microscope (Nikon, Japan, ×200) and the cell numbers were counted in three different fields in every well. All the experiments were repeated in triplicate.
Apoptosis AssayAt 24h post-transfection, cells were trypsinized without ethylene diamine tetraacetic acid (EDTA)and washed using cold phosphate-buffered saline (PBS). FITCAnnexin V and propidium lodide (PI) were added to the cells,then the mixture was incubated for 15min in darkness at room temperature. The cellular apoptosis ratio was analyzed using the Annexin V-FITC apoptosis detection kit (BD Biosciences,San Diego, CA, USA).
RNA Extraction and Real-time Polymerase Chain ReactionTotal RNA from cultured cells was extracted using Trizol reagent (Invitrogen, USA). For miRNA reverse transcription(RT), an miRcute miRNA first-strand cDNA synthesis kit(Tiangen, China) was used. The expression level of miR-22 was quantified using an miRcute plus miRNA-qPCR Detection kit (Tiangen, China) and was normalized to U6 expression.The primer sequences were designed as follows: (U6) forward:5’-GCTTCGGCAGCACATATACTAAAAT-3’; (miR-22) forward: 5’-AAGCTGCCAGTTGAAGAACTGT-3’.The common reverse primer used in this experiment was Uni-miRqRCR. The RT of mRNA was performed with atranscriptor first strand cDNA synthesis kit (Thermo Fisher Scientific, ShangHai, China). SYBR®Premix TaqTM II kit (TaKaRa Bio Inc, Japan) was used to analyze HMGB1 expression, and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz,USA) was designed as the internal control. The primer sequences were designed as follows: (HMGB1) forward:5’-TGTCGGGAGGAGCATAAGA-3’; (HMGB1) reverse:5’-GGGCGATACTCAGAGCAGAA-3’; (GAPDH) forward:5’-GCCGCATCTTCTTTTGCGTC-3’; (GAPDH) reverse:5’-TACGACCAAATCCGTTGACTCC-3’. The RT-PCR assay was conducted on an Applied Biosystems 7500 Real Time PCR system (ABI, CA, USA). The 2-ΔΔCtmethod was used to analyze the relative quantification of HMGB1 and miR-22 expression.
Western BlotWestern blot analysis was performed to detect the expression of endogenous HMGB1, protein kinase B (AKT) and phosphorylated protein kinase B (p-AKT).Following treatment, the whole cell extracts were lysed completely on ice for 20min. Prepared standard sample proteins were loaded onto 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels for electroblotting. And then proteins were transferred onto polyvinylidene fluoride membranes. Add western seal solution (5% skim milk) to the membranes for 2h at room temperature. Proteins were incubated with primary antibodies against HMGB1 (Abcam,UK, ab184211), AKT (CST, USA, 4691) and p-AKT (CST,USA, 4060) at 4℃ overnight, then corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (goat antimouse IgG-HRP, CWBIO, Beijing, China, cw0102s; goat antirabbit IgG-HRP, CWBIO, Beijing, China, cw0103s) were added to samples and incubated for 1h at room temperature.Enhanced chemiluminescence kit (ECL) was used to detect the signals.
Statistical AnalysisSPSS 16.0 software (USA) was used for statistical analysis. Data subject to statistical evaluations are presented as the mean±standard deviation (SD). The differences between two groups were analyzed using the Student’st-test. One-way ANOVA was used when more than two groups were compared.P<0.05 was considered to indicate statistical significance.
RESULTS
Upregulated Expression of miR-22 Inhibited Viability,Migration and Invasion While Promoting Apoptosis in Retinoblastoma Y79 CellsWe constructed a GV251/miR-22 plasmid and transfected it into RB Y79 cells to upregulate the expression of miR-22. As expected, increased miR-22 was observed in the GV251/miR-22 group (Figure 1A). Cell viability in the GV251/miR-22 group was markly decreased according to the CCK-8 assay, with inhibition rate was 11.71%±2.54% (12h),21.36%±1.39% (24h) and 29.44%±1.15% (48h), respectively(Figure 1B). However, cellular apoptosis was higher in the GV251/miR-22 group (17.00%±0.39%) compared with the GV251 group (4.38%±0.38%,P<0.05; Figure 1C, 1D). To subsequently determine the effect of miR-22 on migration and invasion in Y79 cells, scratch and Transwell assays were performed. The cells in the GV251/miR-22 group had a lower migration rate (Figure 1E, 1F). Moreover, over-expression of miR-22 in the cells significantly decreased cell invasion(38.33%±4.11%vs131.00%±3.56%,P<0.05; Figure 1G, 1H).In conclusion, these results suggested that miR-22 promoted cell apoptosis and exhibited an inhibitory effect on viability,migration and invasion in Y79 cells.
miR-22 Negatively Regulated the Expression of Highmobility Group Box 1To understand the mechanism of miR-22 in RB cells, we explored how the expression of HMGB1 was influenced by miR-22. miR-22 inhibited the mRNA and protein levels of HMGB1 in the GV251/miR-22 group (Figure 2A-2C). Furthermore, miR-22 expression was significantly attenuated in the GV249/miR-22-siRNA group compared with that in the control groups (Figure 2D). Moreover, HMGB1 expression was significantly elevated in the GV249/miR-22-siRNA group (Figure 2E-2G). Recent studies validated the hypothesis that miR-22 directly targets the 3’UTR of HMGB1[11,22]. Taken together, these data suggest that miR-22 has a negative regulation on the expression of HMGB1 in Y79 cells.
High-mobility Group Box 1 Silencing by shRNA Inhibited Viability, Migration and Invasion and Promotes ApoptosisNext, we investigate the role of HMGB1 in Y79 cells,functional experiments were performed. Significant decrease of HMGB1 was observed in the GV102/HMGB1-siRNA group(Figure 3A-3C). The 12h post transfection, cell viability in the GV102/HMGB1-siRNA group was significantly decreased in the CCK-8 assay (Figure 3D). Cellular apoptosis was higher in the GV102/HMGB1-siRNA group compared with GV102 group (Figure 3E, 3F). The cells in the GV102/HMGB1-siRNA group exhibited decreased rates of migration (Figure 3G, 3H) and invasion ability (Figure 3I, 3J).
Upregulation of HMGB1 Rescued the Effect of miR-22 Overexpression on Viability, Migration, Invasion and ApoptosisA rescue experiment was conducted to elucidate the functional effects of miR-22 in Y79 cells. Transfected with GV230/HMGB1, Y79 cells exhibited increased expression of HMGB1 (Figure 4A-4C). Y79 cells were then co-transfected with GV251/miR-22 and GV230/HMGB1. The upregulation of HMGB1 partly reversed the effect on viability (after 12h),migration, invasion and apoptosis in Y79 cells induced by miR-22 overexpression (Figure 4D-4J). These data suggested that miR-22 performed biological functions in part by targeting HMGB1 in Y79 cells.

Figure 1 miR-22 over-expression in RB Y79 cells reduced growth, migration and invasion and promoted apoptosis
A: Detection of miR-22 expression in Y79 cells transfected with GV251/miR-22 by RT-PCR; B: A CCK-8 assay demonstrated that transfection with GV251/miR-22 after 12h significantly affected the viability of Y79 cells; C, D: The cellular apoptosis ratio was evaluated using flow cytometry in GV251/miR-22 and control groups; E, F: Determination of migration ability of Y79 cells transfected with GV251/miR-22 by a wound healing assay; G, H:Invasive ability was determined by Transwell assay in GV251/miR-22 and control groups (aP<0.05vsGV251 group).
Biological Functions of miR-22 in Y79 Cells Mediated by the PI3K/AKT Signaling PathwayTo further clarify mechanism underlying the effect of miR-22 in Y79 cells on viability, migration, invasion and apoptosis, the level of p-AKT in the PI3K/AKT signaling pathway was examined. The expression level of p-AKT was significantly decreased in the GV251/miR-22 and GV102/HMGB1-siRNA groups compared with that in normal control group. Furthermore, p-AKT was markly enhanced in the GV230/HMGB1 group. Also, p-AKT was significantly elevated in group co-transfected with GV251/miR-22 and GV230/HMGB1 compared with GV251/miR-22 group (Figure 5A, 5B). The collective results substantiate that miR-22 may serve a functional role in Y79 cells through HMGB1 and the PI3K/AKT signaling pathway may participate in the regulating process.
DISCUSSION
In recent years, accumulating studies have verified that miRNAs serve essential roles in the development of tumors.miRNAs participate in diverse biological functions, including differentiation, proliferation, apoptosis and metastasis[5].Recently, studies have reported that miRNAs can act as both tumor suppressors and tumor promoters[23-24]. miR-22 inhibits the cell motility, invasion and proliferation in human glioblastoma[12]. Poor clinicopathologic characteristics and patient prognosis are associated with low expression of miR-22 in gastric cancer[10]. The prognosis, cell proliferation andtumourigenicity of hepatocellular carcinoma have been correlated with downregulated expression of miR-22[13]. Recent study has shown that miR-22 was low expressed in human RB compared with normal retina[15]. However, the exact role of miR-22 in RB and its molecular mechanisms remain unclear.Our present study examined the effects of miR-22 on the biological characteristics of the RB cell line Y79, through analysis of cell viability, migration, invasion and apoptosis.The present study found that the overexpression of miR-22 inhibited viability, migration and invasion while enhancing apoptosis in Y79 cells. A recent study reported that miR-22-transfected Y79 cells exhibited suppressed proliferation and reduced migration rate[25]. Therefore, miR-22 may function as a tumor suppressor in Y79 cells. As far as we know, this isthe first study to confirm that miR-22 represses invasion and promotes apoptosis in Y79 cells. Similar results have been found in many cancers. For instance, it has been shown that miR-22 is downregulated and inhibits cell migration, invasion and growth in clear renal cell carcinoma[26]. Moreover, a recent study also demonstrated that miR-22 inhibited osteosarcoma cell migration and proliferation[11].
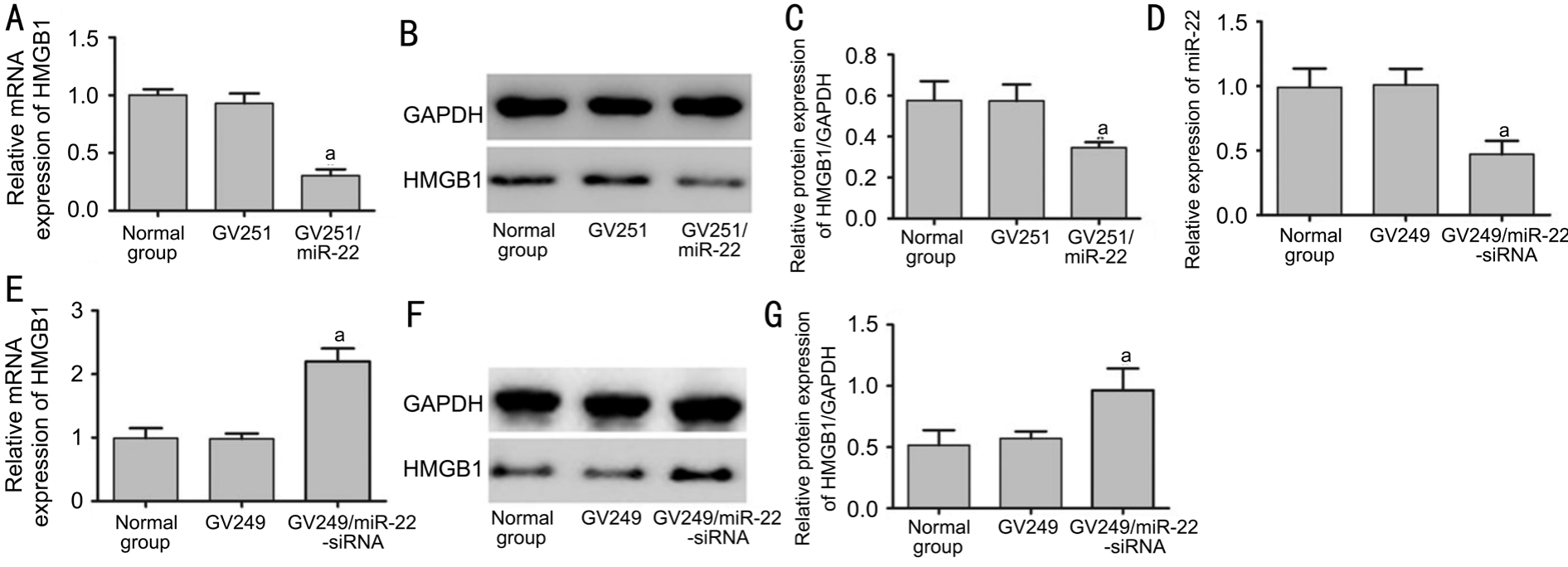
Figure 2 miR-22 negatively regulated HMGB1 expression in RB Y79 cells
A-C: Alterations in HMGB1 in Y79 cells 24h post-transfection GV251/miR-22 were validated by using RT-PCR and Western blot (aP<0.05vsGV251 group); D: Transfection with GV249/miR-22-siRNA significantly downregulated the expression of miR-22 compared with control groups (aP<0.05vsGV249 group); E-G: Alterations in HMGB1 in Y79 cells 24h post-transfection GV249/miR-22-siRNA were validated by using RT-PCR and Western blot (aP<0.05vsGV249 group).
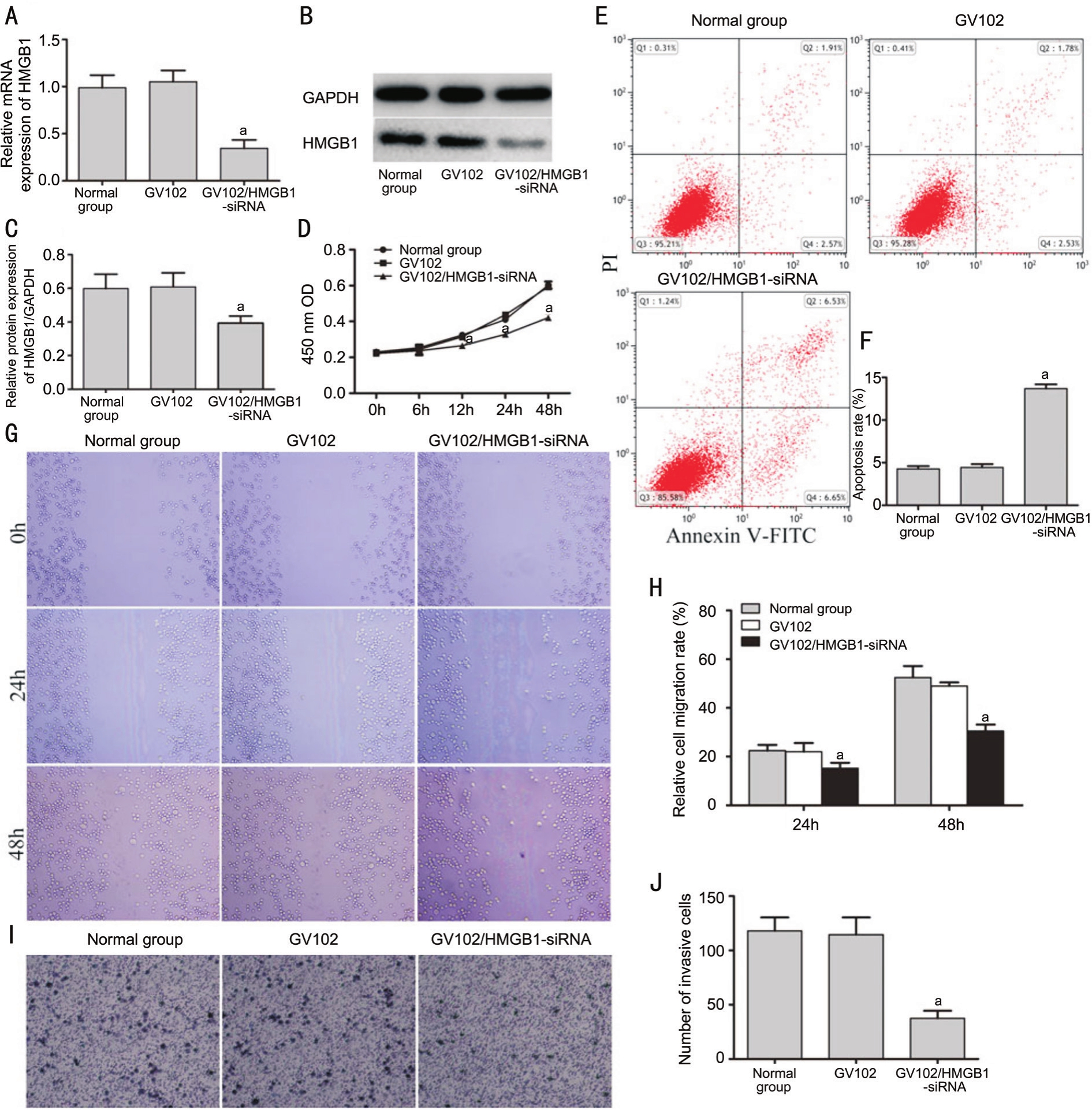
Figure 3 Influence of HMGB1 silencing on growth, migration, invasion and apoptosis in RB Y79 cells
A-C: HMGB1 expression were detected by RT-PCR and Western blot after transfection of Y79 cells with GV102/HMGB1-siRNA or GV102; D: Cell viability was measured by CCK-8 assay in GV102/HMGB1-siRNA and control groups; E, F: The cellular apoptosis ratio was measured by flow cytometry in GV102/HMGB1-siRNA and control groups; G-J: Migratory and invasive abilities were investigated by scratch and Transwell assay in the GV102/HMGB1-siRNA and control groups (aP<0.05vsGV102 group).
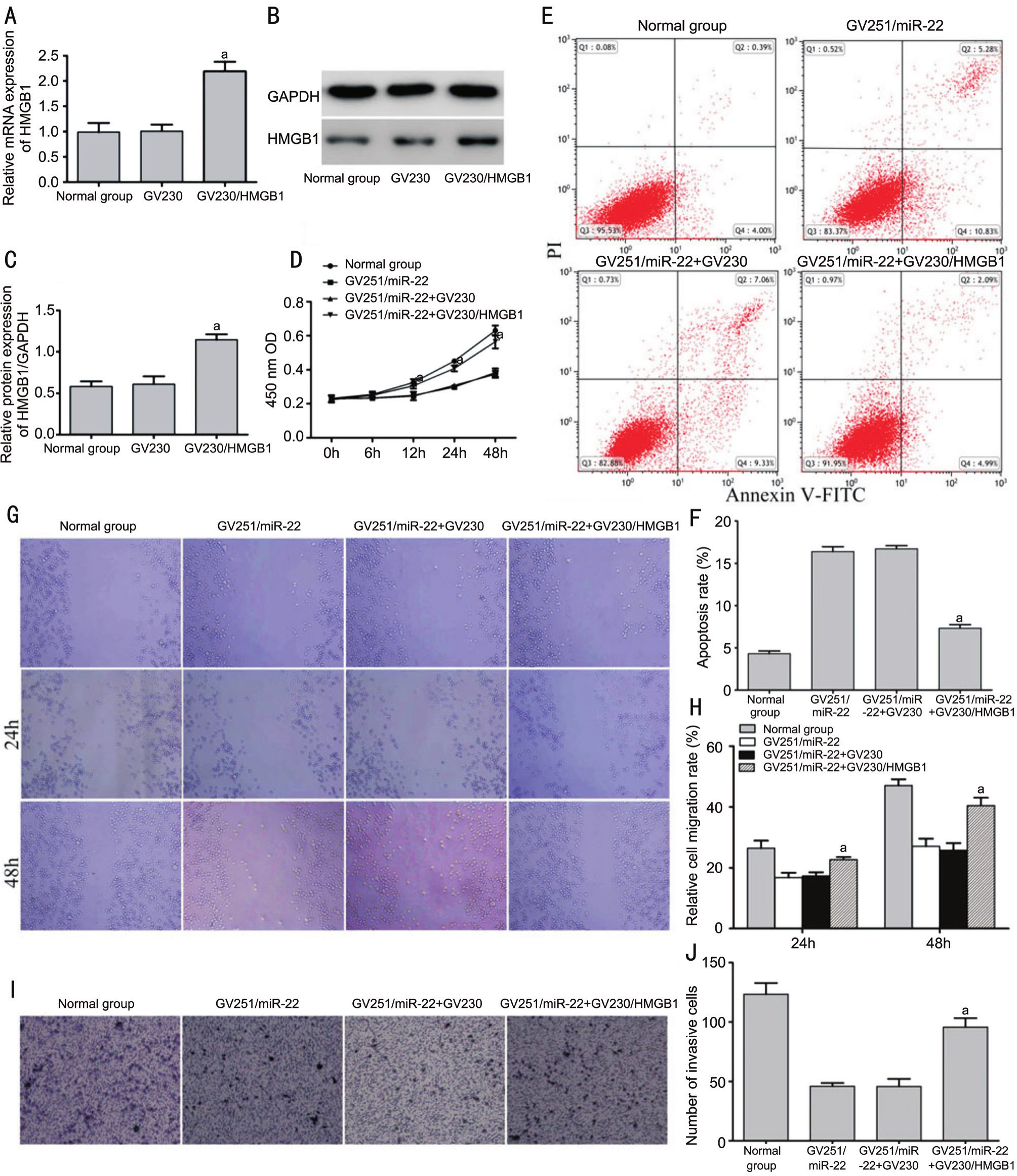
Figure 4 Up-regulation of HMGB1 rescued the effect on proliferation, migration, invasion and apoptosis of miR-22 overexpression in Y79 cells
A-C: The cells were transfected with GV230/HMGB1 and controls. The expression levels of HMGB1 were evaluated by RT-PCR and Western blot (aP<0.05vsGV230 group); D-J: After co-transfection with GV230/HMGB1 and GV251/miR-22, a CCK-8 assay was performed,and cell apoptosis, migration and invasion were investigated (aP<0.05vsGV251/miR-22+GV230 group).
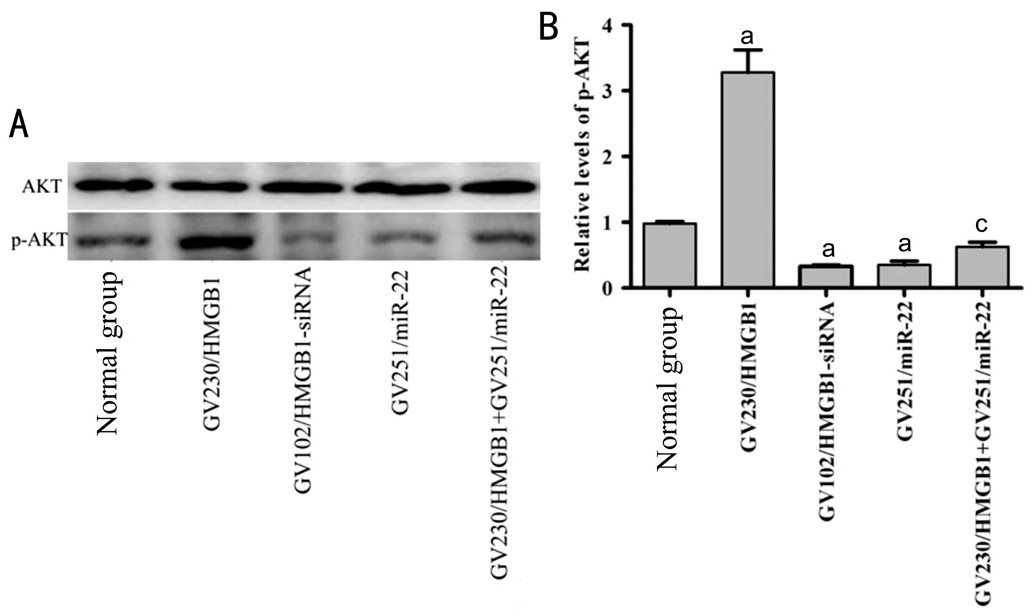
Figure 5 HMGB1 silencing and miR-22 overexpression downregulated p-AKT level in Y79 cells
A, B: The cells were transfected with GV230/HMGB1, GV102/HMGB1-siRNA, GV251/miR-22, GV251/miR-22 and GV230/HMGB1. AKT and p-AKT levels were then measured by Western blot (aP<0.05vsnormal group,cP<0.05vsGV251/miR-22 group).
Recent studies have reported that miR-22 directly targets the 3’-UTR of HMGB1[11,22]. In order to further study the mechanisms of miR-22, the target gene HMGB1 was selected as a candidate gene. First, the association between miR-22 and HMGB1 was investigated in Y79 cells. The data demonstrated that the expression of miR-22 was negatively regulated the expression of HMGB1. Furthermore, viability, migration and invasion were significantly suppressed as a result of decreased HMGB1 expression, while apoptosis was increased. The results were consistent when miR-22 was overexpressed,as miR-22 overexpression downregulated the expression of HMGB1 in RB Y79 cells. Notably, expression of HMGB1 partially rescued the effects of miR-22 overexpression on cell viability, migration, invasion and apoptosis. This suggests that HMGB1, the target gene of miR-22, served an important role in Y79 cells, and that miR-22 may exert biological functions through HMGB1.
A large amount of works have focused on the role of PI3K/AKT signaling in various tumors. Recently, HMGB1 was found to regulate tumor metastasis by activating the PI3K/AKT signaling pathway in cutaneous squamous cell carcinoma[27].Other research has indicated that the PI3K/AKT signaling pathway plays a key role in HMGB1-induced mesangial cell proliferation[28]. To understand the exact mechanism of action of miR-22/HMGB1 in Y79 cells, the p-AKT level in this pathway was assessed. The present study demonstrated that HMGB1 silencing downregulated the degree of p-AKT,while overexpression of HMGB1 enhanced the activation of AKT in Y79 cells. This confirmed that the level of p-AKT was significantly downregulated in the miR-22 overexpression group, and this downregulation of p-AKT induced by miR-22 was partly rescued by HMGB1. These results suggested that miR-22 may exert biological functions on HMGB1 through an additional mechanism; the PI3K/AKT signaling pathway maybe involve in miR-22/HMGB1-induced viability,apoptosis, invasion and migration in Y79 cells.
In conclusion, these results suggested that miR-22 suppressed viability, migration and invasion while promoting apoptosis by targeting HMGB1 in RB Y79 cells. Furthermore, the PI3K/AKT signaling pathway may involve in miR-22-induced changes of biological function. However, the role of miR-22 and HMGB1 in RB Y79 cells needs further investigation.
ACKNOWLEDGEMENTS
Conflicts of Interest:Liu M, None; Wang SM, None; Jiang ZX, None, Lauren H, None; Tao LM, None.
REFERENCES
1 Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS,Gallie BL. Retinoblastoma.Lancet2012;379(9824):1436-1446.
2 Shields CL, Shields JA. Diagnosis and management of retinoblastoma.Cancer Control2004;11(5):317-327.
3 Kivela T. The epidemiological challenge of the most frequent eye cancer:retinoblastoma, an issue of birth and death.Br J Ophthalmol2009;93(9):1129-1131.
4 Dimaras H, Dimba EA, Gallie BL. Challenging the global retinoblastoma survival disparity through a collaborative research effort.Br J Ophthalmol2010;94(11):1415-1416.
5 Bartel DP. MicroRNAs: target recognition and regulatory functions.Cell2009;136(2):215-233.
6 Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight?Nat Rev Genet2008;9(2):102-114.
7 Kagiya T. MicroRNAs and osteolytic bone metastasis: the roles of micrornas in tumor-induced osteoclast differentiation.J Clin Med2015;4(9):1741-1752.
8 Sanei M, Chen X. Mechanisms of microRNA turnover.Curr Opin Plant Biol2015;27:199-206.
9 Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs.Science2001;294(5543):853-858.
10 Wang W, Li F, Zhang Y, Tu Y, Yang Q, Gao X. Reduced expression of miR-22 in gastric cancer is related to clinicopathologic characteristics or patient prognosis.Diagn Pathol2013;8:102.
11 Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang Y, Wang Y, Zhao W,Wang W. miR-22 inhibits osteosarcoma cell proliferation and migration by targeting HMGB1 and inhibiting HMGB1-mediated autophagy.Tumour Biol2014;35(7):7025-7034.
12 Chen H, Lu Q, Fei X, Shen L, Jiang D, Dai D. miR-22 inhibits the proliferation, motility, and invasion of human glioblastoma cells by directly targeting SIRT1.Tumour Biol2016;37(5):6761-6768.
13 Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, Wu M, Pan Z, Zhou W.microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity.Br J Cancer2010;103(8):1215-1220.
14 Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM,Sportoletti P, Varmeh S, Egia A, Fedele G, Rameh L, Loda M, Pandolfi PP. Identification of the miR-106b~25 microRNA cluster as a protooncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation.Sci Signal2010;3(117):ra29.
15 Martin J, Bryar P, Mets M, Weinstein J, Jones A, Martin A, Vanin EF,Scholtens D, Costa FF, Soares MB, Laurie NA. Differentially expressed miRNAs in retinoblastoma.Gene2013;512(2):294-299.
16 Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE,Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, Zeh HJ, Lotze MT.HMGB1 release and redox regulates autophagy and apoptosis in cancer cells.Oncogene2010;29(38):5299-5310.
17 Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1):nuclear weapon in the immune arsenal.Nat Rev Immunol2005;5(4):331-342.
18 Choi YR, Kim H, Kang HJ, Kim NG, Kim JJ, Park KS, Paik YK, Kim HO, Kim H. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation.Cancer Res2003;63(9):2188-2193.
19 Flohr AM, Rogalla P, Meiboom M, Borrmann L, Krohn M, Thode-Halle B, Bullerdiek J. Variation of HMGB1 expression in breast cancer.Anticancer Res2001;21(6A):3881-3885.
20 Poser I, Bosserhoff AK. Transcription factors involved in development and progression of malignant melanoma.Histol Histopathol2004;19(1):173-188.
21 Singh MK, Singh L, Pushker N, Sen S, Sharma A, Chauhan FA,Kashyap S. Correlation of high mobility group box-1 protein (HMGB1)with clinicopathological parameters in primary retinoblastoma.Pathol Oncol Res2015;21(4):1237-1242.
22 Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 targets the 3’ UTR of HMGB1 and inhibits the HMGB1-associated autophagy in osteosarcoma cells during chemotherapy.Tumour Biol2014;35(6):6021-6028.
23 Lujambio A, Lowe SW. The microcosmos of cancer.Nature2012;482(7385):347-355.
24 Xiong J. Emerging roles of microRNA-22 in human disease and normal physiology.Curr Mol Med2012;12(3):247-258.
25 Sreenivasan S, Thirumalai K, Danda R, Krishnakumar S. Effect of curcumin on miRNA expression in human Y79 retinoblastoma cells.Curr Eye Res2012;37(5):421-428.
26 Fan W, Huang J, Xiao H, Liang Z. MicroRNA-22 is downregulated in clear cell renal cell carcinoma, and inhibits cell growth, migration and invasion by targeting PTEN.Mol Med Rep2016;13(6):4800-4806.
27 Sun Y, Tu Y, He LI, Ji C, Cheng BO. High mobility group box 1 regulates tumor metastasis in cutaneous squamous cell carcinoma via the PI3K/AKT and MAPK signaling pathways.Oncol Lett2016;11(1):59-62.
28 Feng X, Wu C, Yang M, Liu Q, Li H, Liu J, Zhang Y, Hao Y, Kang L, Zhang Y, Liu S. Role of PI3K/Akt signal pathway on proliferation of mesangial cell induced by HMGB1.Tissue Cell2016;48(2):121-125.
Citation:Liu M, Wang SM, Jiang ZX, Lauren H, Tao LM. Effects of miR-22 on viability, migration, invasion and apoptosis in retinoblastoma Y79 cells by targeting high-mobility group box 1.Int J Ophthalmol2018;11(10):1600-1607
DOl:10.18240/ijo.2018.10.05
Accepted:2018-07-18
Received:2018-04-23
Correspondence to:Li-Ming Tao. Department of Ophthalmology, the Second Affiliated Hospital of Anhui Medical University, Furong Road, Hefei 230601, Anhui Province,China. taolimingchina@163.com
Foundations:Supported by the Key Project of Anhui Provincial Excellent Young Talent Support Program (No.gxyqZD2017033); the Grant of Second Affiliated Hospital of Anhui Medical University (No.2014BKJ047); the Project of Anhui Provincial Young Wanjiang Scholars Support Program(No.9101041203).




