Genome-wide DNA hypermethylation and homocysteine increase a risk for myopia
Edward Hsi1,2, Yung-Song Wang2,3, Chia-Wei Huang4, Ming-Lung Yu5,6, Suh-Hang Hank Juo1,7,8,9,Chung-Ling Liang10,11
1Centre for Myopia and Eye Disease, Department of Medical Research, China Medical University Hospital, Taichung 404,Taiwan
2Department of Genome Medicine, Kaohsiung Medical University, Kaohsiung 807, Taiwan
3Institute of Fisheries Science, National Taiwan University,Taipei 106, Taiwan
4Department of Biotechnology, Kaohsiung Medical University,Kaohsiung 807, Taiwan
5Hepatobiliary Division, Department of Internal Medicine,Kaohsiung Medical University Hospital, Kaohsiung 807,Taiwan
6Faculty of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 807, Taiwan
7Graduate Institute of Biomedical Sciences, China Medical University, Taichung 404, Taiwan
8Institute of New Drug Development, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan
9Drug Development Center, China Medical University 404,Taiwan
10Department of Ophthalmology, Asia University Hospital,Taichung 413, Taiwan
11Department of Optometry, College of Medical and Health Science, Asia University, Taichung 413, Taiwan
Abstract● AlM: To test for the association between genome-wide methylation and myopia in human and mice.● METHODS: Long interspersed nucleotide element 1 (LINE-1)methylation levels were used to surrogate genome-wide methylation level. We first tested for the association between high myopia (<-6 D) and LINE-1 methylation in leukocytes in 220 cases and 220 control subjects. Secondly, we validated the results of LINE-1 methylation in eyes from the form deprivation myopia (FDM) mice. Furthermore,we calculated the correlation of LINE-1 methylation levels between leukocyte DNA and ocular DNA in the mice. We also tested whether dopamine can alter LINE-1 methylation levels.● RESULTS: The LINE-1 methylation level was significantly higher in the myopic human subjects than controls.The upper and middle tertiles of the methylation levels increased an approximately 2-fold (P≤0.002) risk for myopia than the lower tertile. Similarly, FDM mice had high LINE-1 methylation levels in the leukocyte, retina and sclera, and furthermore the methylation levels detected from these three tissues were significantly correlated.lmmunohistochemical staining revealed higher levels of homocysteine and methionine in the rodent myopic eyes than normal eyes. Dopamine treatment to the cells reduced both LINE-1 methylation and DNA methyltransferase levels.● CONCLUSlON: LINE-1 hypermethylation may be associated with high myopia in human and mice. Homocysteine and methionine are accumulated in myopic eyes, which may provide excess methyl group for genome-wide methylation.
INTRODUCTION
M yopia is a common eye disease worldwide. The prevalence of myopia varies widely among ethnic groups[1-5].Both genetic and environmental factors are important for the development of myopia[6]. Genetic association studies as well as gene expression studies have reported several susceptibility genes to myopia[7-9]. Recent Meta-analysis based on genome-wide association studies further identified several newly identified genetic loci[10-12]. On the other hand, several environmental risk factors were reported to be associated with myopia. Exposure to nearwork and nearwork related parameters such as continuous reading, reading distance, were environmental risk factors for myopia[13]. Outdoor activity is even demonstrated to be a protective factor against myopia onset and progression in school children[14-17]. Data from Metaanalysis also support the possibility of gene X environment interaction as a risk factor for myopia[18].
DNA methylation is a major type of epigenetic regulations,and in general DNA methylation silences gene expression. In adult non-gamete cells, DNA methylation normally occurs at multiple CpG sites in the CpG islands. DNA methylation can be assessed in a global way or gene-specific way. At a global level, the degree of DNA methylation can be measured by utilizing repeat interspersed regions such as long interspersed nucleotide element 1 (LINE-1)[19] because LINE-1 repeats hold approximately 17% of the human genome in size[20]. Several studies measured the LINE-1 methylation levels in leukocyte DNA to test for the association between global methylation and the risk of complex diseases[21-24].
A change of DNA methylation level may result from an interaction between genetic and environmental factors.A global DNA methylation level has been proposed to be influenced by a number of endogenous and environmental factors[25], such as tobacco smoke, air pollution, bisphenol A, and nutrient supplements[26-27]. A major source of methyl donor for DNA methylation originates from homocysteine metabolism, and patients with homocystinuria also have high myopia[28-29]. Homocysteine is biosynthesized from methionine via a multi-step process, and homocysteine can be also recycled into methionine. Previous studies have indicated that administration of homocysteine for a long period time could impair the dopaminergic system[30]. It needs to be noted that intravitreal application of dopamine agonist can suppress the development of myopia in animal studies[31]. Therefore,LINE-1 methylation, homocysteine and dopamine may have a complicated relationship that eventually affects a risk for myopia.
The present study first tested for the association between global LINE-1 methylation level in leukocytes and high myopia in human subjects. We then conducted an animal study to test whether LINE-1 methylation level in leukocytes can reflect LINE-1 status in the ocular tissue. The association between LINE-1 methylation level and myopia was assessed in animals to validate the human study. Homocysteine and methionine were detected in rodent normal and myopic eyes. Finally,cellular studies were conducted to delineate the network connection among dopamine LINE-1 methylation and DNA methyltransferase-1 (DNMT1).
SUBJECTS AND METHODS
Ethical Approval Each subject signed a written informed consent. The study was approved by the Institutional Review Board at the Kaohsiung Medical University Hospital, Taiwan(KMUH-IRB-960238). The research followed the tenets of the Declaration of Helsinki.
The guidelines of animal care are comparable with those published by the Institute for Laboratory Animal Research.The Animal Care and Ethics Committee at Kaohsiung Medical University in Taiwan approved the present animal research(permit No: 103040). The treatment and care of animals were conducted according to the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No.8023, revised 1978).
Study Subject and Sample Size Calculation The participants of present study were recruited from the general population aged between 18 and 30y. A person with previous eye surgery including LASIK surgery was excluded. The participants were enrolled in southern Taiwan between 2003 and 2009. All the participants were of Chinese descent. A case must have myopia in both eyes and the worse eye had a spherical refraction ≤-6.0 diopter (D). A control subject must have a spherical refraction≥-1.5 D and ≤0.75 D in the more myopic eye. Negative cylindrical powers were used in all subjects. Furthermore,a control could not receive any previous refractive surgery.The refractive error was measured without cycloplegia using autorefractometers (Topcon KR-8100 or RM-8800; Topcon,Tokyo, Japan) for all eyes. Based on the data from our previous study of LINE-1 methylation[24], 208 cases and 208 controls would provide a power of 80% with the type I error rate of 0.05.
Pyrosequencing for LINE-1 DNA was isolated by Gentra(Qiagen, Hilden, Germany), according to the manufacturer’s recommendations and stored at -20℃ until used in the subsequent steps. Sodium bisulfate was used to treat DNA samples to convert unmethylated cytosine to uracil, and to leave methylated cytosine intact. An EpiTect Fast Bisul fite Kit(Qiagen) was used for the above mentioned procedure. The completion of bisulfite treatment was evaluated by detecting unconverted bisulfite cytosine outside the CpG assuming that non-CpG cytosines were mainly unmethylated. The biotinylated polymerase chain reaction (PCR) products were purified and pyrosequencing was run on a PyroMark Q24(Qiagen). Non-CpG cytosine residues were used as internal controls to validate the efficiency of sodium bisulfite DNA conversion. Universal unmethylated and methylated DNAs were run as controls. Methylation quantification was performed using the PyroMark Q24 2.0.6 software (Qiagen). The methylation level was calculated as percentage for methylated cytosine over the sum of methylated and unmethylated cytosine.
Induction of Form Deprivation Myopia in Mice The study used C57BL/6J male specific pathogen free mice.These animals were purchased from the National Laboratory Animal Center (Taipei, Taiwan). Mice were maintained in a temperature-controlled (25℃) facility with a strict 12-h light: dark cycle. All animals were provided free access to food and water throughout the experiment. A total of 17 mice (23 days old)were used in this study, among which 6 mice were randomly selected to receive the form deprivation myopia (FDM)and 11 mice were used as normal controls (i.e. free of form deprivation). The right eyes of animals in the FDM group were covered on day 23 for four weeks to induce myopia, while the left eyes were uncovered as previous described[9]. After four weeks, the covers were removed and animals were sacrificed by iso flurane to extract DNA from the blood, retinal and scleral tissues in the laboratory. For FDM mice, only the covered eyes were used for the subsequent experiments.
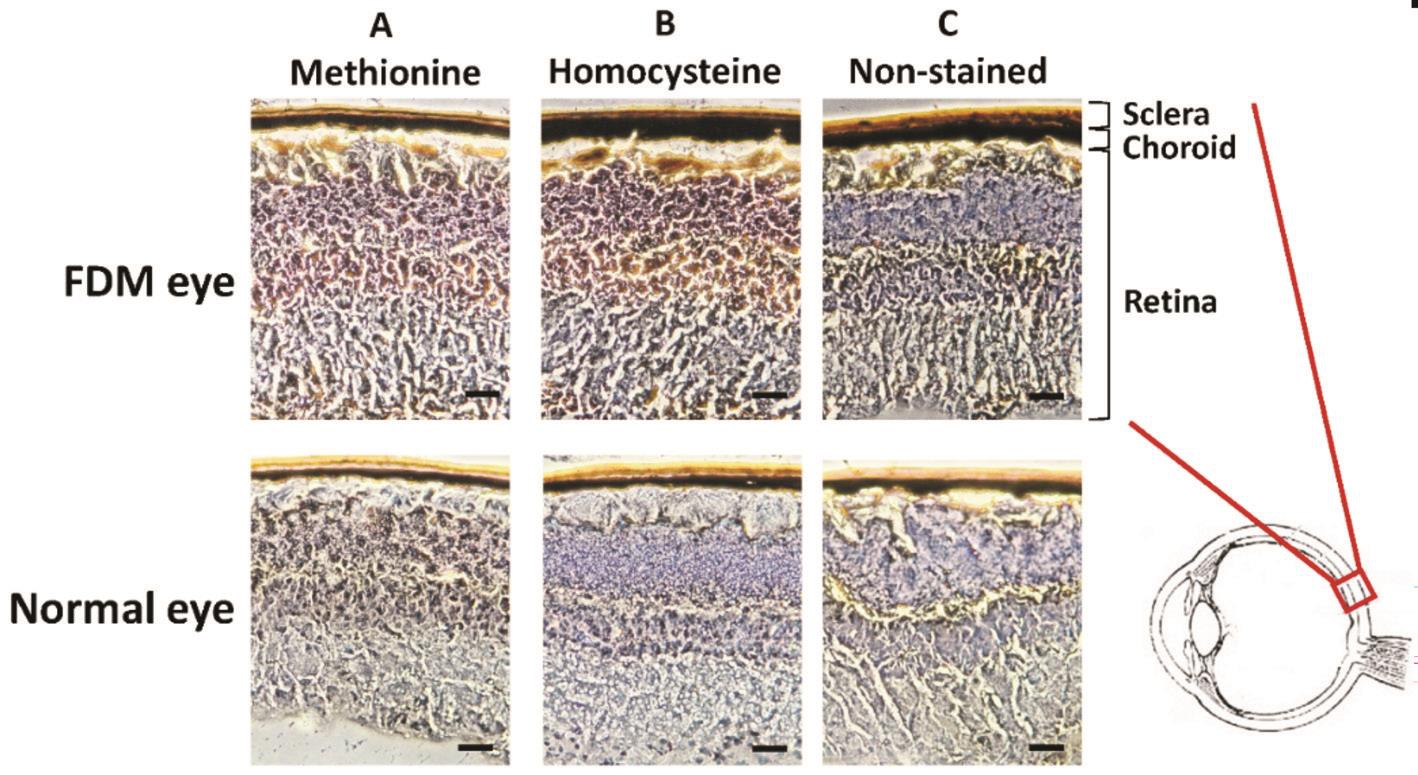
Immunohistochemical Staining for Homocysteine and Methionine Two mice in the FDM group and 2 mice in the control group were randomly selected for the immunocytochemistry study. The eye sections were rinsed twice in PBS and then stained using specific antibodies. The two primary antibodies were mouse anti-methionine (AB6456;1:100 for immunocytochemistry; Abcam, Cambridge,UK) and mouse anti-homocysteine (AB15154; 1:100 for immunocytochemistry; Abcam). These two antibodies were further detected by 0.05% diaminobenzidine (DAB, Vector Laboratories, Burlingame, CA, USA) and 0.03% H2O2. The immunohistochemical (IHC) stained sections were detected by a bright- field microscopy (Leica, Wetzlar, Germany).
Dopamine Treatment to Retinal Pigment Epithelial Cells We purchased the human retinal pigment epithelial(RPE; ARPE-19) cell line from Bioresource Collection and Research Center (BCRC; Hsinchu, Taiwan) which derived from American Type Culture Collection (ATCC, Manassas,VA; ATCC number: CRL-2302). The cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco-BRL,Gaithersburg, MD, USA) supplemented with 2 mmol/L l-glutamine(Sigma-Aldrich, St. Louis, MO, USA), 10% bovine fetal serum (Gibco-BRL), and 1 mmol/L pyruvate (Sigma-Aldrich).The cells were maintained at 37℃ in an atmosphere of 5%CO2. Dopamine (Sigma-Aldrich) was prepared in the final concentration of 10 μmol/L to treat the RPE cells for 24h. The analysis was conducted within five passages of the cells used in the experiments.
RNA Isolation and Quantitative Real-time PCR for DNMT1 Total RNA was extracted using Trizol according to the manufacturer’s instructions and the purity of RNA was checked using A260/A280 readings. cDNA was synthesized from 1 μg total RNA using random primers and the MultiScribe Reverse Transcriptase Kit (Applied Biosystems,Carlsbad, CA, USA). The cDNA was diluted 1:30 with PCR grade water and then stored at -20℃.
For quantitative real-time PCR, specific primers were designed(DNMT1-F: 5’-AGG CGG CTC AAA GA TTT GGA A-3’,DNMT1-R: 5’-GCA GAA ATT CGT GCA AGA GAT TC-3’,GAPDH-F: 5’-GTG AAG GTC GGA GTC AAC-3’,GAPDH-R: 5’-GTT GAG GTC AAT GAA GGG-3’). RNA levels were measured in the ABI 7500 real-time PCR machine(Applied Biosystems) with the pre-optimized condition. Each real-time PCR was performed in triplicate using 1 μL cDNA,0.2 μL primer sets, 5 μL 2×SYBR Green PCR Master Mix,and 3.6 μL nucleotide-free H2O to yield a 10 μL reaction. The expression level of DNMT1 was normalized to that of GAPDH as the internal control.
Statistical Analysis We used the Student’s t test and Chisquare test to analyze the numeric and categorized variables,respectively. We equally divided the total participants into three groups (i.e. tertile) for a post-hoc analysis. Multivariate logistic regression model was used to determine the odds ratio(OR) of each quartile with adjustment for risk factors. The nonparametric Spearman rank correlation was calculated for LINE-1 methylation levels between leukocyte DNA and ocular DNA in the 11 mice. The non-parametric Mann-Whitney test was used to compare LINE-1 methylation levels in animal tissues. A two-sided P value less than 0.05 was considered statistically significant. All statistical calculations were performed by the JMP software (version 9). The statistical power was calculated by G*Power version 3.1.
RESULTS
DNA Methylation in Human Subjects Based on the DNA availability and quality, 220 highly myopic cases and 220 sex- and age-matched controls were used for this study. The demographic data and LINE-1 methylation levels of study subjects are shown in Table 1. The mean refraction error and standard error of mean (SEM) were -8.81±0.14 D for myopic subjects and -0.53±0.04 D for control subjects. The myopic subjects had a higher percentage for a college or higher education level than controls, while sex and age had no significant difference between the cases and controls (Table 1).The LINE-1 methylation level was significantly higher in the myopic subjects than controls (P=0.003, Table 1). We further divided the total population into tertiles. Accordingly, there were 87 cases and 60 controls in the upper tertile, 73 cases and 74 controls in the middle tertile and 60 cases and 86 controls in the lower tertile (Table 1). Most myopic subjects were in the upper (methylation level: 82.23%-91.50%) or middle tertile (methylation level: 79.53%-82.23%), while most control subjects were in the low tertile (methylation level: 70.80%-79.53%). In the multivariate analysis, the OR of high myopia was 2.19 (P=0.001) for the upper tertile and 1.76 (P=0.021) for the middle tertile compared with the lower tertile (Table 1).
Table 1 The association between LINE-1 methylation level and high myopia n (%)
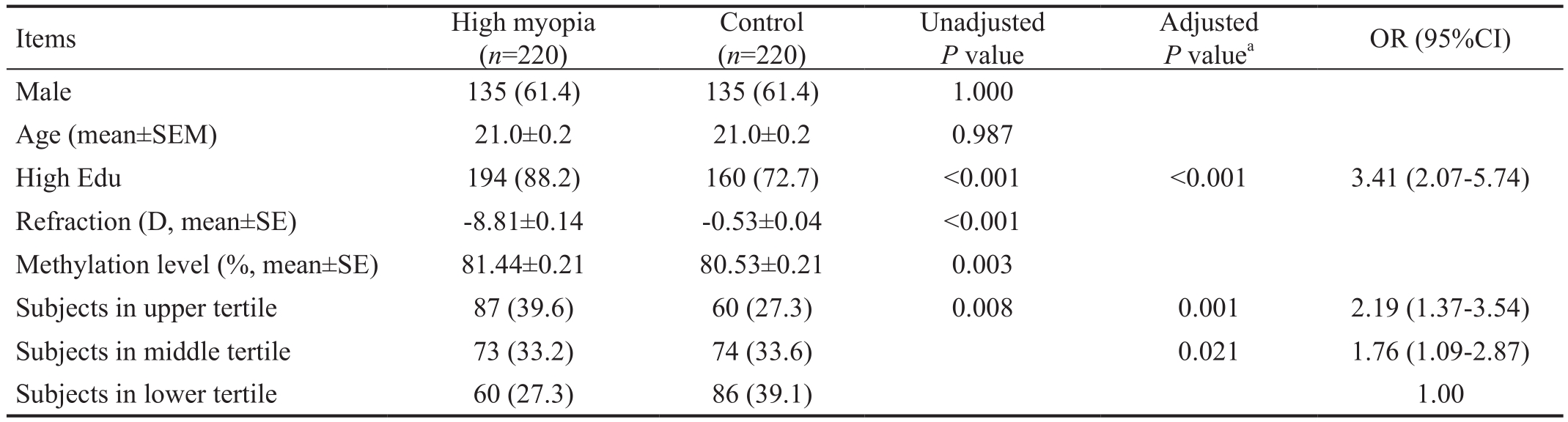
Refraction: Diopter of the worse eye. High myopia: Subjects with refraction error ≤-6 D; Control: Refraction error between 0.75 D and -1.5 D;High Edu: College or higher education level; The range of LINE-1 methylation level (presented as %) for upper title (82.23%-91.50%), middle tertile (79.53%-82.23%) and lower tertile (70.80%-79.53%).aAdjusted for sex, age and education.
Items High myopia(n=220)Control(n=220)Unadjusted P value Adjusted P valuea OR (95%CI)Male 135 (61.4) 135 (61.4) 1.000 Age (mean±SEM) 21.0±0.2 21.0±0.2 0.987 High Edu 194 (88.2) 160 (72.7) <0.001 <0.001 3.41 (2.07-5.74)Refraction (D, mean±SE) -8.81±0.14 -0.53±0.04 <0.001 Methylation level (%, mean±SE) 81.44±0.21 80.53±0.21 0.003 Subjects in upper tertile 87 (39.6) 60 (27.3) 0.008 0.001 2.19 (1.37-3.54)Subjects in middle tertile 73 (33.2) 74 (33.6) 0.021 1.76 (1.09-2.87)Subjects in lower tertile 60 (27.3) 86 (39.1) 1.00
DNA Methylation in Experimental Animals Although measurement of methylation in leukocyte DNA is practically feasible, there may be a concern about the consistent change of methylation between DNA of leukocyte and ocular tissues, including retina and sclera. In our animal model,the rodent LINE-1 methylation level in the peripheral blood was significantly correlated with that in the ocular tissue in 11 normal mice (Figure 1). The correlation coefficients of methylation levels were 0.63 (P=0.038) between the retina and leukocyte, and 0.68 (P=0.020) between the sclera and leukocyte. Furthermore, LINE-1 methylation levels in leukocyte, sclera and retina were significantly higher in the FDM mice than normal mice with a P value of 0.027, 0.018 and 0.032, respectively (Figure 2). The statistic power was greater than 0.80. Such data not only confirm the association between LINE-1 methylation level and myopia, but also justify the use of leukocyte DNA to surrogate ocular DNA for methylation studies in myopia.
Immunohistochemical Staining for Homocysteine and Methionine Because homocysteine metabolism provides the methyl donor for DNA methylation, we then examined homocysteine and methionine expression in the mouse eyes.Four mice (two from the FDM group and two from the control group) that were originally used for methylation study were randomly selected for the IHC staining. The representative images are shown in Figure 3. The results showed that the retina of FDM had increased staining of homocysteine and methionine than the retina from the normal mice. There was no observable difference of IHC staining for either homocysteine or methionine in the sclera between FDM and normal eyes(Figure 3). The above animal study suggested that FDM eyes had a higher homocysteine metabolism, which is consistent with human data[28-29].
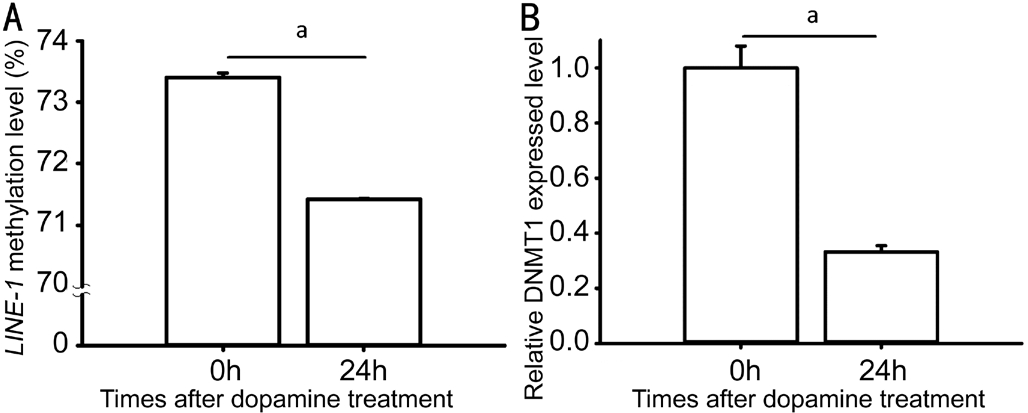
Dopamine Reduced LINE-1 Methylation Level There is no consensus on which cell type is the best for in vitro myopia studies. Because the above IHC data showed increased homocysteine in the retina, we decided to use a convenient human RPE cell to conduct the subsequent cell study.Dopamine has been repeatedly reported to effectively reduce myopia development in experimental animals[31]. To explore whether dopamine affects methylation process, we measured LINE-1 methylation and DNMT1 levels in the dopaminetreated RPE cells. Because DNMT1 is the most abundant DNA methyltransferase in mammalian cells, it is likely to be the key maintenance methyltransferase in mammals. After the 24h dopamine treatment, both LINE-1 methylation level and DNMT1 RNA level were significantly reduced (P=0.030 and 0.001, respectively) in the RPE cells (Figure 4).
DISCUSSION
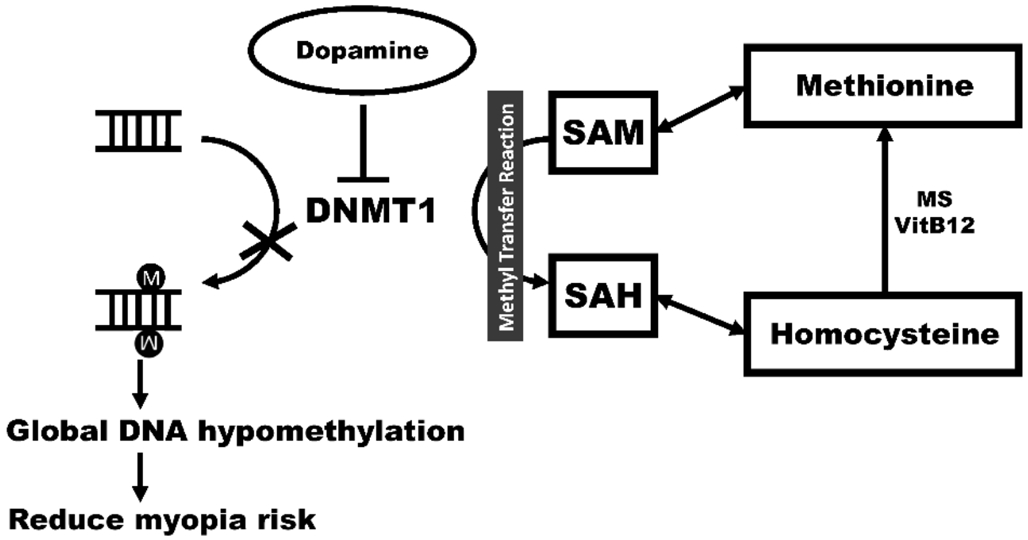
Our study demonstrated that elevated global DNA methylation levels of LINE-1 in the leukocytes were independently associated with high myopia in human subjects. In the mouse study, we showed that the LINE-1 methylation levels in leukocytes and ocular tissue were significantly correlated. This result suggests peripheral blood may surrogate the ocular tissue in terms of LINE-1 methylation status. Our further animal experiment revealed that DNA isolated from rodent leukocyte,retina and sclera had higher LINE-1 methylation levels in the myopic mice. Therefore, the animal studies justify the validity of measuring LINE-1 status in peripheral leukocytes and confirm the result of human study. The concentrations of two amino acids (methionine and homocysteine) involved in the methylation process were higher in myopic eyes than normal eyes. Such data suggest excessive methyl donor in myopic eye leading to LINE-1 hypermethylation. Our study also first ever demonstrated that dopamine can decrease DNMT1, which in turn reduces LINE-1 methylation level. The present study provides an epigenetic mechanism for dopamine therapy in experimental myopia. Our finding and possible molecular mechanism is schematically depicted in Figure 5.
Homocysteine can be methylated and then be converted to methionine (Figure 5). Methionine is an essential amino acid and can be activated to S-adenosylmethionine (SAM) that is a methyl donor for several metabolic reactions including DNA methylation. After losing its methyl group for DNA methylation, SAM is converted to S-adenosylhomocysteine(SAH) that can be further metabolized to homocysteine. Then,methionine can be regenerated from homocysteine. Myopic subjects have been reported to have high levels of circulating methionine[32]. Furthermore, homocystinuria is characterized by the presence of severe myopia[29,33], which implies accumulated homocysteine is a risk factor for myopia. Excessive methionine and homocysteine suggest a plenty of methyl group for LINE-1 DNA methylation, which may contribute to myopia development. In addition, a positive correlation between circulating methionine and leukocyte LINE-1 methylation was also found in colorectal cancer patients[34].
Dopamine plays an important role in the vertebrate visual system, and a reduction of retinal dopamine results in decreased visual contrast sensitivity[35]. Injection of dopamine agonists to the eyes can inhibit FDM development in animal models while injection of dopamine antagonists can enhance FDM development[31]. In addition, dopamine antagonist impairs the effect of bright light on myopia prevention[36]. Here we demonstrated that dopamine could reduce DNMT1 that is an enzyme for DNA methylation. Consistently, we found that dopamine could lower the LINE-1 methylation level.These findings may provide another mechanism to explain the dopamine effect on inhibiting myopia development.
Aberrant DNA methylation has been implied in several complex diseases[37-40]. Gene-environment interaction has long been considered a risk for myopia, but the molecule evidence to support this speculation is relatively sparse. Although DNA methylation can be a marker to test for gene-environment interaction, only few related studies in the context of myopia have been reported. Recently, Zhou et al[41] demonstrated that COL1A1 expression is modulated by DNA methylation during experimental myopia in mice. In their study, the methylation level in the COL1A1 promoter increased in the sclera of FDM eyes, but the level remained low in the fellow eye.Consequently, the FDM eyes had lower COL1A1 expression levels than the fellow and normal eyes. Discovery of the involvement of DNA methylation in myopia development not only provides more insight to emmotropization but also implies a potential opportunity for myopia intervention.
The study design has strengths and limitations. The study population was relatively homogenous in respect to age,ethnicity and geographic location. There may be a concern of misclassification of our control subjects if their refraction still progresses. However, it is well known that refraction progression slows down with age and mild myopia is unlikely to progress after age of 18[42-43]. Even though few control subjects had myopia progression to make them illegible as controls, our results should remain significant given that the original data had a P value of 0.003. However, the cross-sectional association study cannot test for the causal relationship. Therefore, we used the animal study to offer another line of evidence for LINE-1 hypermethylation as a risk for myopia. Another limitation is that the present study does not explain the exact role of LINE-1 in the myopia development. Since LINE-1 biological role has not been extensively studied, our results should be used as evidence to support the role of DNA methylation in myopia development.Further studies to disclose aberrant DNA methylation in specific genes are warranted to gain more understanding in myopia pathogenesis.
Another concern is the measure of refraction without cycloplegia. A previous study using the subjects of similar age(aged 22 to 39y) reported that refraction measured without cycloplegia can be over-estimated by -0.23 D in myopes(defined as -1 D or less) and -0.43 D for emmetropes (defined as between -1 D to +1 D)[44]. Another study (aged 18 to 34y)reported that refraction without cycloplegia caused a more myopic estimate by -0.86 D regardless of the myopic or emmetropic participants[45]. However, both studies showed that the over-estimation is smaller for myopes than emmetropes.Since the mean of refraction is -8.81 D for myopic subjects and -0.53 D for our controls, the non-cycloplegia effect would not change their disease status and would not affect the results and conclusion.
In conclusion, the present study demonstrates that the methylation level in leukocyte LINE-1 is associated with myopia in both human and mice. Rodent LINE-1 methylation levels measured in blood and ocular tissue are significantly correlated, which justifies the measure of methylation in peripheral blood for myopia studies. Furthermore, our results indicate that dopamine can reduce both LINE-1 methylation and DNMT1 levels, which provides a novel mechanism for dopamine effect on myopia prevention.
ACKNOWLEDGEMENTS
Authors’ contributions: Hsi E designed and conducted the study, analyzed the data and wrote the manuscript; Wang YS designed and conducted the study; Huang CW conducted the study; Yu ML designed the study and interpreted the data; Juo SH designed the study, analyzed and interpreted the data and wrote the manuscript; Liang CL designed the study, interpreted the data and wrote and approved the manuscript.
Foundations: Supported by the grants from the Taiwan Ministry of Science and Technology (MOST 104-2622-B-037-005); Taiwan Ministry of Health and Welfare Clinical Trial Centre (MOHW107-TDU-B-212-123004) and Drug Development Center, China Medical University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Confiicts of Interest: Hsi E, None; Wang YS, None; Huang CW, None; Yu ML, None; Juo SH, None; Liang CL, None.
REFERENCES
1 Wu HM, Seet B, Yap EP, Saw SM, Lim TH, Chia KS. Does education explain ethnic differences in myopia prevalence? a population-based study of young adult males in Singapore. Optom Vis Sci 2001;78(4):234-239.
2 Pan CW, Chen Q, Sheng X, Li J, Niu Z, Zhou H, Wei T, Yuan Y, Zhong H.Ethnic variations in myopia and ocular biometry among adults in a rural community in China: the Yunnan minority eye studies. Invest Ophthalmol Vis Sci 2015;56(5):3235-3241.
3 Lin LL, Shih YF, Hsiao CK, Chen CJ, Lee LA, Hung PT. Epidemiologic study of the prevalence and severity of myopia among schoolchildren in Taiwan in 2000. J Formos Med Assoc 2001;100(10):684-691.
4 Li SM, Liu LR, Li SY, et al. Design, methodology and baseline data of a school-based cohort study in Central China: the Anyang Childhood Eye Study. Ophthalmic Epidemiol 2013;20(6):348-359.
5 Katz J, Tielsch JM, Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci 1997;38(2):334-340.
6 Liang CL, Yen E, Su JY, Liu C, Chang TY, Park N, Wu MJ, Lee S,Flynn JT, Juo SH. Impact of family history of high myopia on level and onset of myopia. Invest Ophthalmol Vis Sci 2004;45(10):3446-3452.
7 Liang CL, Hsi E, Chen KC, Pan YR, Wang YS, Juo SH. A functional polymorphism at 3’UTR of the PAX6 gene may confer risk for extreme myopia in the Chinese. Invest Ophthalmol Vis Sci 2011;52(6):3500-3505.
8 Liang CL, Wang HS, Hung KS, Hsi E, Sun A, Kuo YH, Juo SH.Evaluation of MMP3 and TIMP1 as candidate genes for high myopia in young Taiwanese men. Am J Ophthalmol 2006;142(3):518-520.
9 Hsi E, Chen KC, Chang WS, Yu ML, Liang CL, Juo SH. A functional polymorphism at the FGF10 gene is associated with extreme myopia.Invest Ophthalmol Vis Sci 2013;54(5):3265-3271.
10 Verhoeven VJ, Hysi PG, Wojciechowski R, et al. Genome-wide metaanalyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet 2013;45(3):314-318.
11 Simpson CL, Wojciechowski R, Oexle K, et al. Genome-wide metaanalysis of myopia and hyperopia provides evidence for replication of 11 loci. PLoS One 2014;9(9):e107110.
12 Miyake M, Yamashiro K, Tabara Y, et al. Identification of myopiaassociated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. Nat Commun 2015;6:6689.
13 Li SM, Li SY, Kang MT, et al. Near work related parameters and myopia in Chinese children: the Anyang Childhood Eye Study. PLoS One 2015;10(8):e0134514.
14 Guggenheim JA, Northstone K, McMahon G, Ness AR, Deere K,Mattocks C, Pourcain BS, Williams C. Time outdoors and physical activity as predictors of incident myopia in childhood: a prospective cohort study. Invest Ophthalmol Vis Sci 2012;53(6):2856-2865.
15 Jones LA, Sinnott LT, Mutti DO, Mitchell GL, Moeschberger ML,Zadnik K. Parental history of myopia, sports and outdoor activities, and future myopia. Invest Ophthalmol Vis Sci 2007;48(8):3524-3532.
16 Li SM, Li H, Li SY, et al. Time outdoors and myopia progression over 2 years in Chinese children: the Anyang Childhood Eye Study. Invest Ophthalmol Vis Sci 2015;56(8):4734-4740.
17 Wu PC, Tsai CL, Wu HL, Yang YH, Kuo HK. Outdoor activity during class recess reduces myopia onset and progression in school children.Ophthalmology 2013;120(5):1080-1085.
18 Fan Q, Verhoeven VJ, Wojciechowski R, et al. Meta-analysis of geneenvironment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun 2016;7:11008.
19 Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E,Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005;33(21):6823-6836.
20 Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009;10(10):691-703.
21 Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L,Sparrow D, Vokonas P, Schwartz J. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 2010;21(6):819-828.
22 Muka T, Koromani F, Portilla E, O’Connor A, Bramer WM, Troup J, Chowdhury R, Dehghan A, Franco OH. The role of epigenetic modifications in cardiovascular disease: a systematic review. Int J Cardiol 2016;212:174-183.
23 Chen D, Zhang XR, Zhang Y, Zhang L, Ma JL, You WC, Pan KF.Hypomethylation of repetitive elements in blood leukocyte DNA and risk of gastric lesions in a Chinese population. Cancer Epidemiol 2016;41:122-128.
24 Lin RT, Hsi E, Lin HF, Liao YC, Wang YS, Juo SH. LINE-1 methylation is associated with an increased risk of ischemic stroke in men. Curr Neurovasc Res 2014;11(1):4-9.
25 Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33(Suppl):245-254.
26 Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2012;13(2):97-109.
27 Schernhammer ES, Giovannucci E, Kawasaki T, Rosner B, Fuchs CS,Ogino S. Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut 2010;59(6):794-799.
28 Karaca M, Hismi B, Ozgul RK, Karaca S, Yilmaz DY, Coskun T,Sivri HS, Tokatli A, Dursun A. High prevalence of cerebral venous sinus thrombosis (CVST) as presentation of cystathionine beta-synthase deficiency in childhood: molecular and clinical findings of Turkish probands. Gene 2014;534(2):197-203.
29 Sacharow SJ, Picker JD, Levy HL. Homocystinuria caused by cystathionine beta-synthase deficiency. Genereviews (Internet). May 18, 2017.
30 Lee ES, Chen H, Soliman KF, Charlton CG. Effects of homocysteine on the dopaminergic system and behavior in rodents. Neurotoxicology 2005;26(3):361-371.
31 Feldkaemper M, Schaeffel F. An updated view on the role of dopamine in myopia. Exp Eye Res 2013;114:106-119.
32 Wuu JA, Wen LY, Chuang TY, Chang GG. Amino acid concentrations in serum and aqueous humor from subjects with extreme myopia or senile cataract. Clin Chem 1988;34(8):1610-1613.
33 Yap S, Naughten E. Homocystinuria due to cystathionine beta-synthase deficiency in Ireland: 25 years’ experience of a newborn screened and treated population with reference to clinical outcome and biochemical control. J Inherit Metab Dis 1998;21(7):738-747.
34 Jung AY, Botma A, Lute C, Blom HJ, Ueland PM, Kvalheim G,Midttun Ø, Nagengast F, Steegenga W, Kampman E. Plasma B vitamins and LINE-1 DNA methylation in leukocytes of patients with a history of colorectal adenomas. Mol Nutr Food Res 2013;57(4):698-708.
35 Witkovsky P. Dopamine and retinal function. Doc Ophthalmol 2004;108(1):17-39.
36 Ashby RS, Schaeffel F. The effect of bright light on lens compensation in chicks. Invest Ophthalmol Vis Sci 2010;51(10):5247-5253.
37 Wu H, Zhao M, Tan L, Lu Q. The key culprit in the pathogenesis of systemic lupus erythematosus: Aberrant DNA methylation. Autoimmun Rev 2016;15(7):684-689.
38 Lin HF, Hsi E, Liao YC, Chhor B, Hung J, Juo SH, Lin RT.Demethylation of circulating estrogen receptor alpha gene in cerebral ischemic stroke. PLoS One 2015;10(9):e0139608.
39 Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human cancer. Oncotarget 2016;7(24):37331-37346.
40 Wu H, Zhao M, Chang C, Lu Q. The real culprit in systemic lupus erythematosus: abnormal epigenetic regulation. Int J Mol Sci 2015;16(5):11013-11033.
41 Zhou X, Ji F, An J, et al. Experimental murine myopia induces collagen type Ialpha1 (COL1A1) DNA methylation and altered COL1A1 messenger RNA expression in sclera. Mol Vis 2012;18:1312-1324.
42 Pärssinen O, Kauppinen M. What is the in fluence of parents’ myopia on their children’s myopic progression? A 22-year follow-up study. Acta Ophthalmol 2016;94(6):579-585.
43 Sayegh FN. Age and refraction in 46,000 patients as a potential predictor of refractive stability after refractive surgery. J Refract Surg 2009;25(8):747-751.
44 Krantz EM, Cruickshanks KJ, Klein BE, Klein R, Huang GH, Nieto FJ. Measuring refraction in adults in epidemiological studies. Arch Ophthalmol 2010;128(1):88-92.
45 Jorge J, Queiros A, González-Méijome J, Fernandes P, Almeida JB, Parafita MA. The influence of cycloplegia in objective refraction.Ophthalmic Physiol Opt 2005;25(4):340-345.
Citation: Hsi E, Wang YS, Huang CW, Yu ML, Juo SH, Liang CL.Genome-wide DNA hypermethylation and homocysteine increase a risk for myopia. Int J Ophthalmol 2019;12(1):38-45
DOl:10.18240/ijo.2019.01.06
● KEYWORDS: methylation; myopia; LINE-1; homocysteine;dopamine
Received: 2018-05-25 Accepted: 2018-11-27
Correspondence to: Suh-Hang Hank Juo. China Medical University, Taichung, 91 Hsueh-Shih Road, Taichung 404,Taiwan. hjuo@mail.cmu.edu.tw; Chung-Ling Liang. Asian University Hospital, Department of Ophthalmology, 500 Lioufeng Road, Taichung City 41354, Taiwan. ling0228@gmail.com