
Citation: Zhang YH, Xing YQ, Chen Z, Ma XC, Lu Q. Association
between interleukin-10 genetic polymorphisms and risk of primary open angle
glaucoma in a Chinese Han population: a case-control study. Int J Ophthalmol 2019;12(10):1605-1611. DOI:10.18240/ijo.2019.10.13
·Investigation·
Association
between interleukin-10 genetic polymorphisms and risk of primary open angle
glaucoma in a Chinese Han population: a case-control study
Yi-Hui Zhang1,2, Yi-Qiao Xing1,
Zhen Chen1, Xiao-Cheng Ma2, Qiang Lu2
1Department of Ophthalmology, Renmin
Hospital of Wuhan University, Wuhan 430060, Hubei Pronvince, China
2Ophthalmology Department of Inner
Mongolia People’s Hospital, Hohhot 010017, Inner Mongolia Autonomous Region,
China
Correspondence to: Yi-Qiao Xing. Department of
Ophthalmology, Renmin Hospital of Wuhan University, 99 Zhangzhidong Road,
Wuchang, Wuhan 430060, Hubei Pronvince, China. drxingyiqiao@163.com
Received:
Abstract
AIM: To investigate the association between interleukin-10 (IL-10) genetic
polymorphisms and risk of POAG through a case-control study in a Han population
of China.
METHODS: A total of 210 patients with POAG and 420 normal
subjects were recruited during the period from Dec. 2013 to Dec. 2016. The
IL-10
RESULTS: We observed that those carrying the CC genotype of
rs1800871 was associated with an increased risk of POAG when compared with
those harboring the TT genotype (OR=1.84, 95%CI=1.01-3.38). Those with AA
genotype of rs1800872 had a 10.62 fold risk of POAG in comparison to the CC
genotype (OR=10.62, 95%CI, 3.41-33.09). A completely linkage disequilibrium was
found between IL-10 rs1800871-rs1800872 (D’=1.00, r2=0.16).
The A-C-A (OR=2.60, 95%CI, 1.48-4.58) and G-T-A (OR=2.34, 95%CI, 1.42-3.86)
haplotypes were associated with an increased risk of POAG, while the A-T-C
haplotype showed a decreased risk of POAG (OR=0.63, 95%CI, 0.49-0.81).
CONCLUSION: Our data suggest that IL-10 rs1800871 and rs1800872
can be predictive factors for the pathogenesis of POAG in the Chinese
population.
KEYWORDS: primary open angle glaucoma; IL-10;
polymorphism; haplotype
DOI:10.18240/ijo.2019.10.13
Citation:
Zhang YH, Xing YQ, Chen Z, Ma XC, Lu Q. Association between interleukin-10
genetic polymorphisms and risk of primary open angle glaucoma in a Chinese Han
population: a case-control study. Int J Ophthalmol 2019;12(10):1605-1611
INTRODUCTION
Glaucoma is the leading cause of
irreversible blindness[1], and this disease has
became one of the important public health issues worldwide[2].
It is estimated that about 60.5 million people with primary glaucoma by 2010,
increasing to 79.6 million by 2020 with bilateral blindness in 5.9 million
primary open angle glaucoma (POAG)[3]. Almost half
of the world’s glaucoma population occur in Asian counties. A recent study
suggests that POAG prevalence is about 0.7% in mainland China[4].
The etiology of developing POAG is still uncertain, but it is well known that
many environmental and lifestyle factors contribute to the development of this
disease, such as intraocular pressure, age, alcohol drinking, cigarette
smoking, high body mass index, systemic hypertension[5].
However, individuals with similar risk factors of POAG would not develop this
disease, suggesting that genetic factors play an important role in the
pathogenesis of POAG. Currently, many studies have indicated many genetic
factors contribute to the development of POAG, such as MTHFR C677T, vitamin D,
GSTM1, OPA1, CYP
Studies have linked inflammation and
immune reaction play critical roles in the processes of POAG[12-13]. Interleukin-10 (IL-10), also known as human cytokine
synthesis inhibitory factor (CSIF), is a multifunctional cytokine which is
mainly come from human T helper type1 (Th1) cells and Th2 cells, macrophages/monocytes
and dendritic cells[14]. IL-10 exhibits complex
immunosuppressive and immunostimulatory properties. For instance, IL-10
promotes B cell-mediated functions, enhancing proliferation, differentiation,
and antibody production, although it can inhibit the functions of T cells and
antigen presenting cells (APCs), host type 1 immune responses were inhibited by
reducing the production of IL-2, IFN-gamma, and other cytokines, IL-10 itself
inhibits the production of IFN-gamma from Th1 cells, and the lack of IFN-gamma
increases APC inactivation[15]. Further research
revealing more about important roles of IL
SUBJECTS AND METHODS
Ethical Approval A case-control study design was
used. In this study, 210 patients with POAG without any blood relationship were
recruited from the Renmin Hospital of Wuhan University between December 2013
and December 2016. Informed written consent was obtained from all participants
for use of their blood sample for genotyping for this study. The study protocol
was approved by the Institutional Review Board of Renmin Hospital of Wuhan
University, China (No.201601005).
Subjects All the patients were diagnosed by
the following criteria: 1) abnormal appearance of the disc or retinal nerve
fiber layer; 2) visual field loss according to optic nerve damage; 3)
glaucomatous optic nerve damage and cup-to-disc ratio (CDR) above 0.5; 4)
intraocular pressure above
Simultaneously, a total of 420
controls were recruited from the individuals attending the routine health
examination and with no previous history of glaucoma. The patients and controls
were genetically unrelated Chinese Han population.
DNA Extraction Totally 5 mL of peripheral venous blood
were obtained from each respondent. The white cell was handled with Qiagen
Blood DNA extraction kit to extract genomic DNA for genotyping analysis
according to the manufacturer’s protocol, and stored at
IL-10 Genotyping The IL-10
Statistical Analysis Demographic and clinical
characteristics, and genotype and allele frequencies of the IL-10
The association between IL-10
RESULTS
The demographic and clinical
variables of 210 patients with POAG and 420 healthy controls are shown in Table
1. No significant difference was found between patients with POAG and controls
in terms of age (χ2=0.76, P=0.86), sex (χ2=0.003,
P=0.96), BMI (χ2=0.02, P=0.89) and smoking
status (χ2=0.03, P=0.86). However, we found that
patients with POAG were more likely to have a history of POAG (χ2=11.84,
P=0.001), diabetes (χ2=9.84, P=0.002) and
hypertension (χ2=10.86, P=0.001), a habit of drinking
(χ2=6.30, P=0.01), and higher levels of intraocular
pressure (t=44.68, P<0.001) and CDR (t=49.19, P<0.001).
Table 1 Demographic and clinical
characteristics of patients with POAG and healthy controls n (%)
|
Variables |
Patients (n=210) |
Controls (n=420) |
χ2 or t |
P |
|
Age, y |
|
|
0.76 |
0.86 |
|
<45 |
33 (15.71) |
64 (15.24) |
|
|
|
45-60 |
57 (27.14) |
128 (30.48) |
|
|
|
60-75 |
85 (40.48) |
162 (38.57) |
|
|
|
>75 |
35 (16.67) |
66 (15.71) |
|
|
|
Sex |
|
|
0.003 |
0.96 |
|
Female |
94 (44.76) |
187 (44.52) |
|
|
|
Male |
116 (55.24) |
233 (55.48) |
|
|
|
BMI, kg/m2 |
|
|
0.02 |
0.89 |
|
<24 |
163 (77.62) |
328 (78.10) |
|
|
|
≥24 |
47 (22.38) |
92 (21.90) |
|
|
|
History of POAG |
|
|
11.84 |
0.001 |
|
No |
201 (95.71) |
418 (99.52) |
|
|
|
Yes |
9 (4.29) |
2 (0.48) |
|
|
|
History of diabetes |
|
|
9.84 |
0.002 |
|
No |
175 (83.33) |
385 (91.67) |
|
|
|
Yes |
35 (16.67) |
35 (8.33) |
|
|
|
History of hypertension |
|
|
10.86 |
0.001 |
|
No |
146 (69.52) |
341 (81.19) |
|
|
|
Yes |
64 (30.48) |
79 (18.81) |
|
|
|
Smoking |
|
|
0.03 |
0.86 |
|
Never |
131 (62.38) |
265 (63.10) |
|
|
|
Ever |
79 (37.62) |
155 (36.90) |
|
|
|
Drinking |
|
|
6.30 |
0.01 |
|
Never |
118 (56.19) |
279 (66.43) |
|
|
|
Ever |
92 (43.81) |
141 (33.57) |
|
|
|
Intraocular pressure, mm Hg |
26.51±2.27 |
15.93±2.93 |
44.68 |
<0.001 |
|
Cup-to-disk ratio |
0.75±0.11 |
0.34±0.09 |
49.19 |
<0.001 |
We observed that the TT, TC, and CC
genotypes of rs1800871 showed significantly differences between patients with
POAG and controls (χ2=6.19, P=0.04), and the CC, CA
and AA genotypes of rs1800872 also revealed a significantly differences (χ2=28.81,
P<0.001; Table 2). Moreover, we found that the rs1800870, rs1800871
and rs1800872 were in line with Hardy-Weinberg equilibrium in both patients and
controls.
Table 2 Genotype distributions of
IL-10
|
SNPs |
Patients (n=210) |
Controls (n=420) |
χ2 |
P value |
Patients |
Controls |
||
|
χ2 for HWE |
P for HWE |
χ2 for HWE |
P for HWE |
|||||
|
rs1800870 |
|
|
3.55 |
0.17 |
0.21 |
0.56 |
0.34 |
0.56 |
|
AA |
82 (39.05) |
197 (46.90) |
|
|
|
|
|
|
|
AG |
105 (50.00) |
185 (44.05) |
|
|
|
|
|
|
|
GG |
23 (10.95) |
38 (9.05) |
|
|
|
|
|
|
|
rs1800871 |
|
|
6.19 |
0.04 |
0.05 |
0.82 |
0.42 |
0.39 |
|
TT |
85 (40.48) |
178 (42.38) |
|
|
|
|
|
|
|
TC |
96 (45.71) |
210 (50.00) |
|
|
|
|
|
|
|
CC |
29 (13.81) |
32 (7.62) |
|
|
|
|
|
|
|
rs1800872 |
|
|
28.81 |
<0.001 |
3.42 |
0.08 |
0.012 |
0.91 |
|
CC |
142 (67.62) |
340 (80.95) |
|
|
|
|
|
|
|
CA |
50 (23.81) |
76 (18.10) |
|
|
|
|
|
|
|
AA |
18 (8.57) |
4 (0.95) |
|
|
|
|
|
|
Using conditional logistic
regression analysis, we observed that those carrying the CC genotype of
rs1800871 were associated with an increased risk of POAG when compared with
those harboring the TT genotype (OR: 1.84, 95%CI: 1.01-3.38). Those with AA
genotype of rs1800872 had a 10.62 fold risk of POAG in comparison to the CC
genotype (OR: 10.62, 95%CI: 3.41-33.09; Table 3).
Table 3 Logistic regression analysis
of association between IL-10 polymorphisms and risk of POAG patients
|
Variables |
β |
S.E. |
Wals |
OR |
95%CI |
P |
|
rs1800870 |
|
|
|
|
|
|
|
AA |
|
|
|
|
1.0 (Ref.) |
- |
|
AG |
0.28 |
0.19 |
2.04 |
1.32 |
0.90-1.92 |
0.153 |
|
GG |
0.12 |
0.33 |
0.13 |
1.12 |
0.59-2.13 |
0.719 |
|
rs1800871 |
|
|
|
|
|
|
|
TT |
|
|
|
|
1.0 (Ref.) |
- |
|
TC |
0.20 |
0.20 |
1.05 |
1.22 |
0.83-1.80 |
0.306 |
|
CC |
0.61 |
0.31 |
3.91 |
1.84 |
1.01-3.38 |
0.048 |
|
rs1800872 |
|
|
|
|
|
|
|
CC |
|
|
|
|
1.0 (Ref.) |
- |
|
CA |
0.35 |
0.22 |
2.48 |
1.41 |
0.92-2.17 |
0.115 |
|
AA |
2.36 |
0.58 |
16.62 |
10.62 |
3.41-33.09 |
<0.001 |
|
History of POAG |
|
|
|
|
|
|
|
No |
|
|
|
|
1.0 (Ref.) |
- |
|
Yes |
2.27 |
0.81 |
7.92 |
9.65 |
1.99-46.79 |
0.005 |
|
History of diabetes |
|
|
|
|
|
|
|
No |
|
|
|
|
1.0 (Ref.) |
- |
|
Yes |
0.68 |
0.28 |
6.03 |
1.98 |
1.15-3.43 |
0.014 |
|
History of hypertension |
|
|
|
|
|
|
|
No |
|
|
|
|
1.0 (Ref.) |
- |
|
Yes |
0.66 |
0.21 |
10.32 |
1.93 |
1.29-2.89 |
0.001 |
|
Drinking habit |
|
|
|
|
|
|
|
Never |
|
|
|
|
1.0 (Ref.) |
- |
|
Ever |
0.38 |
0.19 |
4.12 |
1.46 |
1.01-2.09 |
0.042 |
In addition, we found that
individuals with a history of POAG (OR: 9.65, 95%CI: 1.99-46.79), diabetes (OR:
1.98, 95%CI: 1.15-3.43) and hypertension (OR: 1.93, 95%: 1.29-2.89), and a
habit of drinking status (OR: 1.46, 95%CI: 1.01-2.09) had an increased risk of
POAG, when compared with the reference group.
A completely linkage disequilibrium
was found between IL-10 rs1800871 and rs1800872 (D’: 1.00, r2:
0.16; Figures 1 and 2). The A-C-A (OR: 2.60, 95%CI: 1.48-4.58) and G-T-A (OR:
2.34, 95%CI: 1.42-3.86) haplotypes were associated with an increased risk of
POAG (Table 4), while the A-T-C haplotype showed a decreased risk of POAG (OR:
0.63, 95%CI: 0.49-0.81).
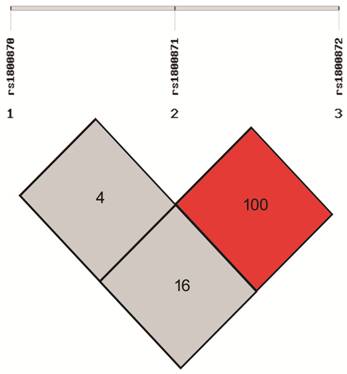
Figure 1 D’ of linkage
disequilibrium test for IL-10 rs1800870, rs1800871, and rs1800872.
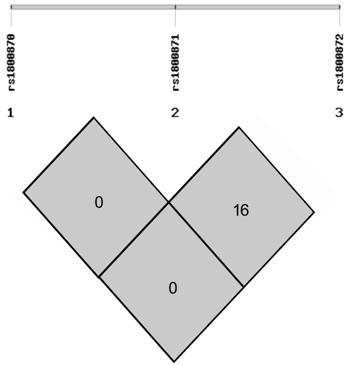
Figure 2 r2 of
linkage disequilibrium test for IL-10 rs1800870, rs1800871, and rs1800872.
Table 4 Haplotype analysis of IL-10
rs1800870, rs1800871, and rs1800872 with POAG risk n (%)
|
Haplotype |
Patients |
Controls |
OR (95%CI) |
P |
|
A-C-A |
28 (6.67) |
23 (2.74) |
2.60 (1.48-4.58) |
<0.001 |
|
A-C-C |
82 (19.52) |
154 (18.33) |
1.12
(0.83-1.51) |
0.47 |
|
A-T-C |
141 (33.57) |
377 (44.88) |
0.63 (0.49-0.81) |
<0.001 |
|
G-C-C |
38 (9.05) |
93 (11.07) |
0.83
(0.56-1.24) |
0.36 |
|
G-T-A |
34 (8.10) |
31 (3.69) |
2.34
(1.42-3.86) |
<0.001 |
|
G-T-C |
73 (17.38) |
132 (15.71) |
1.16
(0.84-1.59) |
0.36 |
DISCUSSION
A misbalance in the physiological
equilibrium may shift from regulatory immunity into a neuroinflammatory
degenerative process, what may lead to a predisposition to glaucoma. As POAG
has been characterized as a neurodegenerative disorder which similar to other
neurodegenerative diseases, increasing research concerning the role of the
immune system in POAG has been performed in the recent years that demonstrates
the immune system definitely plays a role in the pathogenesis of POAG[23-25].
Among the many immune cytokines,
IL-10 exhibits a double-faced role during the development of cancers and
inflammation related diseases, inducing both immunosuppressive and
anti-angiogenic effect. Previous studies revealed that cytokine gene
polymorphisms contributed to the development of ocular involvement and many eye
related diseases[26-29]. It is
reported that early acute inflammatory condition occurs in eye with current
acute primary angle-closure, and anti-inflammatory treatment could be a useful
method for acute primary angle-closure[30]. POAG
is correlated with an aqueous inflammatory response in the aqueous humor, and
the inflammatory response is significantly elevated in eyes[31-32]. Therefore, expression of IL-10 may be associated
with the pathogenesis of POAG. Another hypothesis has gained strength in recent
years, variants SNPs in the population may contribute significantly to genetic
risk for common diseases including age-related disorders. It is well-known for
a long time that many primary eye diseases, including POAG, have genetic
components. Polymorphisms of the related genes of POAG, have been shown to have
some role in the development of glaucoma[33-36].
In our study, we found that the
IL-10 rs1800871 and rs1800872 were significant associated with an increased
risk of POAG, and a completely linkage disequilibrium was found between IL-10
rs1800871 and rs1800872.
SNPs which play an important role in
the regulation the expression of protein, can contribute to the differences
between individuals in the susceptibility to a disease and its severity. The
human IL-10 gene is located on chromosome 1q31-1q32 and composed of five exons
and four introns[20]. In the IL-10 gene promoter
region, the alleles of
There are three limitations should
be mentioned in the present study. First, since patients and controls were only
enrolled from one place of China, these participants may not well represent
individuals in other places, and the selection bias may be inevitable. Second,
since the incidence of POAG was low, the sample size of patients was relatively
small, which may result in a low statistical power to identify differences
between groups. Third, this is a case-control study, which could not explain
the causal relationship between risk factors and diseases.
In conclusion, our data suggest that
IL-10 rs1800871 and rs1800872 could be considered as a predictive factor for
the pathogenesis of POAG in the Chinese population.
ACKNOWLEDGEMENTS
Conflicts of Interest: Zhang YH, None; Xing YQ, None;
Chen Z, None; Ma XC, None; Lu Q, None.
REFERENCES