Conjunctival microbiome changes associated with fungal keratitis: metagenomic analysis
Cheng Ge1,2, Chao Wei2, Bao-Xia Yang2, Jun Cheng2, Yu-Sen Huang2
1Department of Medicine, Qingdao University, Qingdao 266071, Shandong Province, China
2State Key Laboratory Cultivation Base, Shandong Provincial Key Laboratory of Ophthalmology, Shandong Eye Institute,Shandong Academy of Medical Sciences, Qingdao 266071,Shandong Province, China
Abstract● AlM: To investigate the ocular surface microbiome profile of patients with fungal keratitis (FK) through bacterial 16S rDNA sequencing.● METHODS: The swab samples were collected from 8 patients with FK (Group 1 from the corneal ulcer, Group 2 from the conjunctival sac of the infected eyes, and Group 3 from the conjunctival sac of the fellow eyes) and 10 healthy eyes (Group 4 from the conjunctival sac). Bacterial 16S rDNA V4-V5 region sequencing was performed to characterize the bacterial communities on the ocular surfaces of the patients with FK.● RESULTS: Our metagenomic data showed that 97% of the sequence reads were categorized into 245 distinct bacterial genera, with 67.75±7.79 genera detected in Group 1, 73.80±13.44 in Group 2, 74.57±14.14 in Group 3, and 89.60±27.49 in Group 4. Compared with the healthy eyes(Group 4), both infected (Groups 1 and 2) and fellow eyes (Group 3) of the patients with FK showed reduced bacterial diversity and altered ocular surface microbiota compositions, with lower abundance of Corynebacterium and Staphylococcus and higher abundances of Pseudomonas, Achromobacter, Caulobacter and Psychrobacter.● CONCLUSlON: Our report depicts the altered ocular surface bacterial community structures both in the affected and fellow eyes of patients with FK. These changes may contribute to the pathogenesis of FK or the increased risk for FK.
INTRODUCTION
F ungal keratitis (FK) is a kind of severe corneal infectious disease and remains challenging worldwide because of many obstacles to the effective prevention, diagnosis and therapy. Once the initial anti-fungal treatment is delayed,patients have to receive keratoplasty surgery, sometimes with a poor prognosis. Corneal trauma is the most common risk of FK[1], but there are many FK patients without any history of trauma. Undefirstanding of the epidemiology and pathophysiologic mechanism of FK needs to be improved.A growing body of evidence suggests that the human ocular surface is colonized by diverse commensal microbial communities, which play important roles in homeostatic maintenance of healthy eyes[2-5]. Dysbiosis of the ocular surface also drives the pathogenesis of ocular diseases[3,6-8]. The normal microbiota has a protective immunoregulatory function against pathogenic species, and alterations of the normal microbiome may lead to the disease[3,9-10]. Clinical data have also proved that the abuse of topical antibiotics correlates positively with fungal infection[11]. Moreover, Corynebacterium as an ocular commensal has been found to modulate ocular immunity and protect the ocular surface from fungal infection in mice[12].These findings suggest the involvement of ocular surface microbiota into the pathogenesis of FK. Therefore, it is necessary to investigate the ocular surface microbiome profile of patients suffering FK.
Recent microbiological studies are supported technically by metagenomics, a combination of genomics, bioinformatics,and system biology[13], which is the development of highthroughput DNA sequencing and bioinformatics technology.We previously investigated the composition and diversity of bacterial flora in the normal conjunctiva and identified the pathogenic bacteria from aqueous humor or vitreous body in the eyes with endophthalmitis by using 16S rDNA sequencing technique[14-15]. In the current study, we characterized the bacterial communities on the ocular surface of patients with FK using high-throughput 16S rDNA sequencing metagenomic analysis, finding that the altered commensal bacteria of ocular surface may contribute to the pathogenesis of FK or the increased risk for FK.
SUBJECTS AND METHODS
Ethical Approval The Ethics Committee of Shandong Eye Institute approved the prospective, observational study. Written informed consent was obtained from all the studied subjects for sample collection and subsequent analyses. The investigations and measurements were performed in accordance with the Declaration of Helsinki.
Sample Collection Eight patients with FK and 10 controls without ocular surface disease who were hospitalized between March 1 and April 30, 2015 were included in the study. All participants did not have any systemic medical administration within 6mo and underwent a complete ophthalmic examination. FK was diagnosed based on the corneal scraping revealing fungal elements in smears with 10% potassium hydroxide (KOH), culture and confocal microscopy. All patients with FK developed monocular disease and their fellow eyes did not receive any eyedrops. The 10 controls were patients scheduled for cataract surgery, who did not have a medical history of systemic disease or receive eyedrops administration (antibiotics, corticosteroids, and nonsteroidal anti-inflammatory agents) within 6mo. Eight corneal scraping samples from infected eyes (Group 1) were obtained with a sterile surgical blade following the instillation of 0.5%tetracaine hydrochloride eyedrops into the eye. Conjunctival specimens from affected eyes (Group 2) and fellow eyes(Group 3) of patients with FK, and control eyes (Group 4)were collected from the upper, lower palpebral conjunctiva,caruncle and fornix conjunctiva under topical anesthesia using disposable sterile dry absorbent cotton swabs. Both the corneal scrapings and cotton swabs were placed in sterile tubes and stored in a -80℃ freezer until DNA extraction.
DNA ExtractionDNA from different samples was extracted using the MicroElute Genomic DNA Kit (D3096-01; Omega,Guangzhou, China) according to the manufacturer's instructions.Sample blanks consisting of unused swabs were processed through DNA extraction for veri fication of no DNA. The total DNA of swabs was eluted in 20 mL of elution buffer (Omega D3096) and stored at -80℃ until measurement by polymerase chain reaction (PCR; Lianchuan Biotech, Hangzhou, China).
PCR Amplification and 16S rDNA Sequencing Using the total DNA from the 34 samples as a template and the primer (319F 5'-ACTCCTACGGGAGGCAGCAG-3'; 806R 5'-GGACTACHVGGGTWTCTAAT-3'), we amplified the V3-V4 regions of the bacterial 16S rDNA. All reactions were carried out in 25 mL (total volume) of mixtures containing approximately 25 ng of genomic DNA extract, 12.5 mL of PCR premix, 2.5 mL of each primer, and PCR grade water to adjust the volume. PCR reactions were performed in the Master Cycler gradient thermocycler (Eppendorf, Hamburg,Germany), including initial denaturation at 98℃ for 30s, 35 cycles of denaturation at 98℃ for 10s, annealing at 54℃/52℃for 30s, extension at 72℃ for 45s, and final extension at 72℃for 10min. The PCR products were confirmed with 2% agarose gel electrophoresis. During the DNA extraction process,ultrapure water, instead of a sample solution, was used to exclude any false positive PCR results. The PCR products were normalized by the AxyPrep Mag PCR Normalizer (Axygen Biosciences, Union City, CA, USA), which could skip the quantification step despite the PCR volume submitted for sequencing. The amplicon pools were prepared for sequencing with AMPure XT beads (Beckman Coulter Genomics,Danvers, MA, USA). The size and quantity of the amplicon library were assessed with both the LabChip GX (PerkinElmer,Waltham, MA, USA) and the Library Quantification Kit for illumina (Kapa Biosciences, Woburn, MA, USA). PhiX Control library (Illumina) was combined with the amplicon library (expected at 30%) and sequenced on 300PE MiSeq runs using the standard illumina sequencing primers.
Bioinformatics Analysis Samples were sequenced on an illumina MiSeq platform (LC-Bio) according to the manufacturer's instructions. Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequence. After the pairedend reads were merged using the FLASH (Maryland, USA)[16],quality filtering on the raw tags were performed under specific filtering conditions to obtain the high-quality clean tags according to the FastQC (V 0.10.1; Cambridge, UK). Chimeric sequences were filtered using the Verseach software (v2.3.4)(Norway). Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs) by Verseach(v2.3.4; Norway). Representative sequences were chosen for each OTU, and taxonomic data were then assigned to each representative sequence using the Ribosomal Database Project classifier. Multiple sequence alignments were conducted using the PyNAST software (Boulder, USA) to study the phylogenetic relationship of different OTUs. OTUs abundance information was normalized using a standard sequence number corresponding to the sample with the least sequences. Alpha diversity was employed to analyze complexity of species diversity for each sample through 4 indices, including Chao1,Shannon, Simpson, and Observed species, calculated with the QIIME (Version 1.8.0; Boulder, USA). Beta diversity calculated by the principle co-ordinates analysis (PCoA) and cluster analysis using the QIIME software (Version 1.8.0;Boulder, USA) was used to evaluate differences of samples in species complexity. The taxonomy and abundance were bioinformatically analyzed at phylum, class, order, family,genus, and species levels.
Statistical Analysis All statistical analyses were performed using the R (Version 3.2.5; New Zealand) and GraphPad prism 5.0 (GraphPad Software, San Diego, CA, USA). Alpha index and the relative abundance of the four groups were compared by the one-way analysis of variance, and the difference between any two groups was compared by the Kruskal-Wallis test. The results are expressed as mean±standard deviations(SD) and a P value of <0.05 was considered statistically significant.
RESULTS
Operational Taxonomic Units Analysis The involved patients were 10 men and 8 women, with an average age of 58.3±9.53y (range, 41 to 78). Among the 34 subjects, 30 samples (30/34) with sufficient quantity gene amplicons were used for OTU analysis.
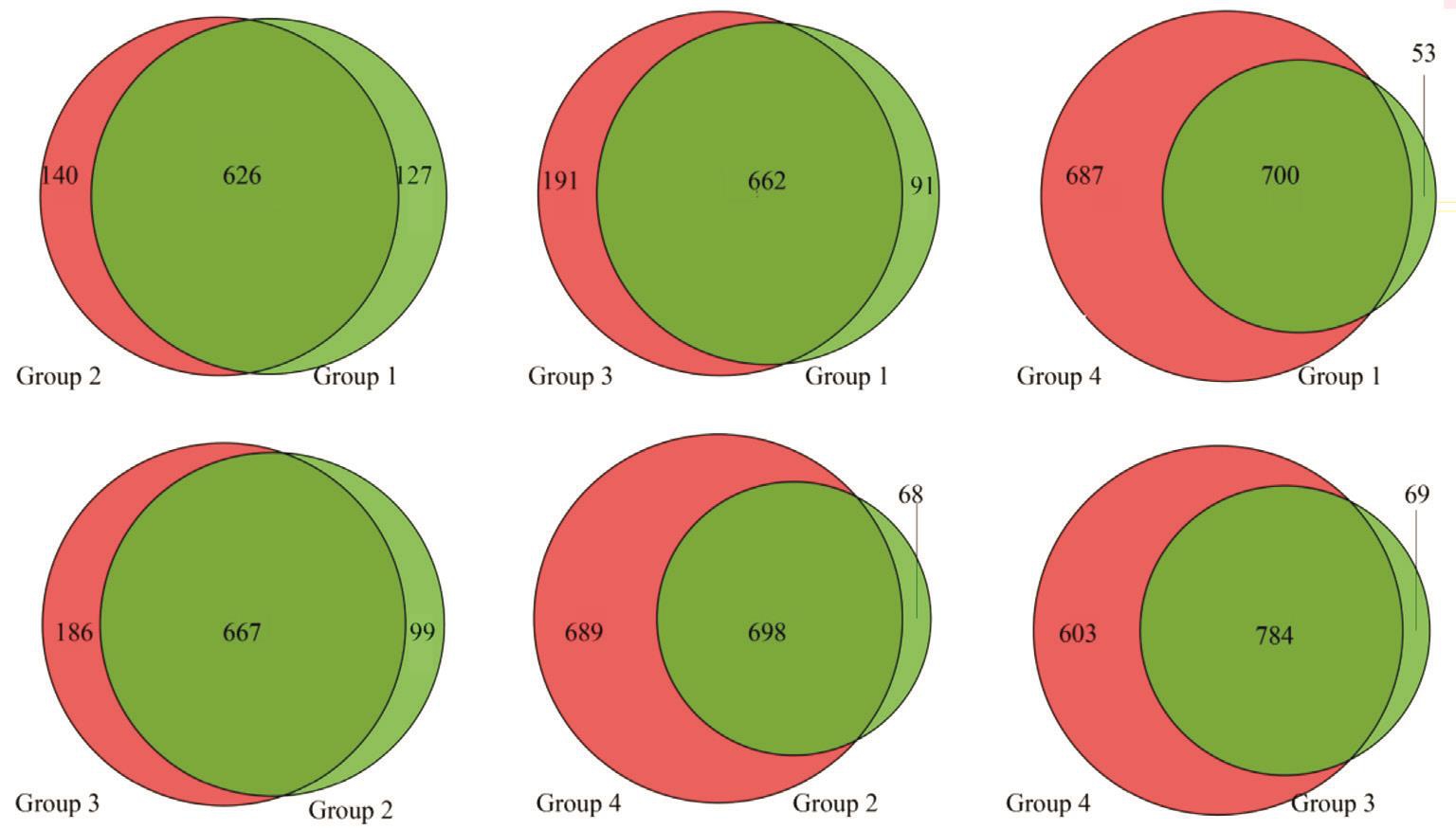
Totally 750 900 high-quality sequencing reads were obtained from 30 samples after the removal of low-quality reads.The OTU classification was performed on the high-quality sequences using Verseach software (v2.3.4, Norway). At the OTU level, the difference between different groups was compared, and both the common and the unique OTUs between different groups were shown through the Venn diagram (Figure 1).
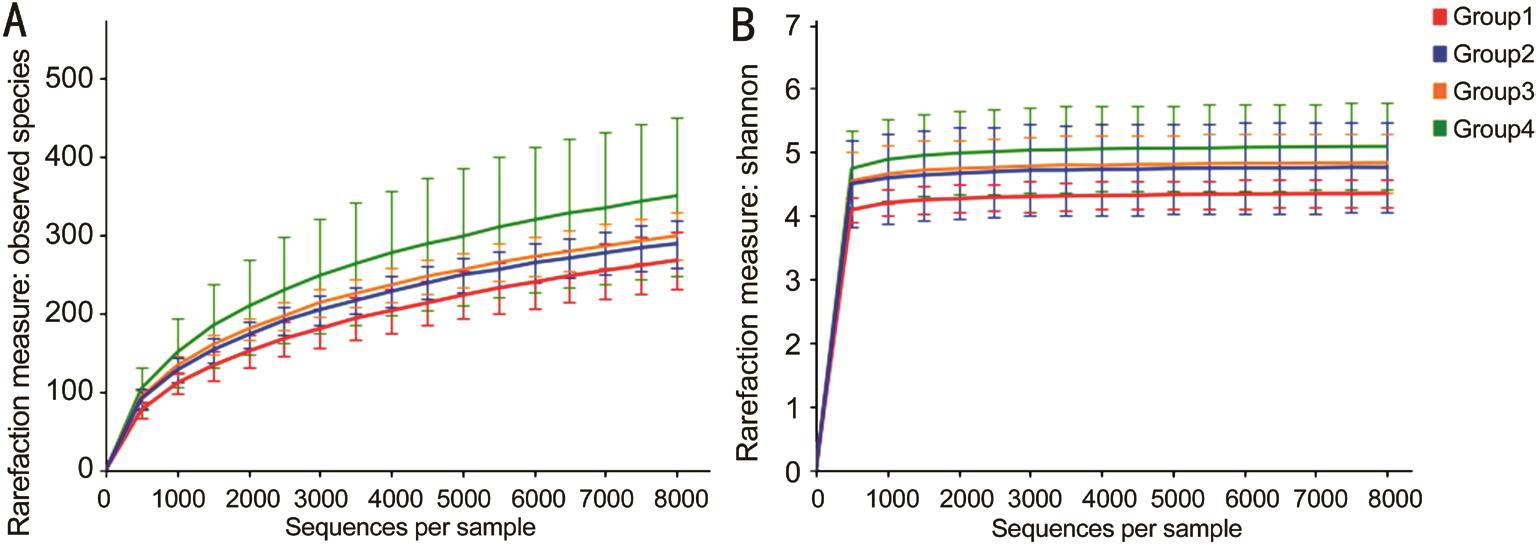
Alpha Diversity Analysis The diversity of microbial communities in the subjects was assessed with alpha diversity analysis. The observed species and Shannon index were used to evaluate the richness and biodiversity of the microbiota(Figure 2). Comparison of the bacteria in the four groups revealed similar trajectories despite the varied number of reads.The four groups had diverse bacterial communities, showing that the samples from FK patients (Groups 1, 2 and 3) were relatively less diverse than those from the healthy eyes (Group 4).
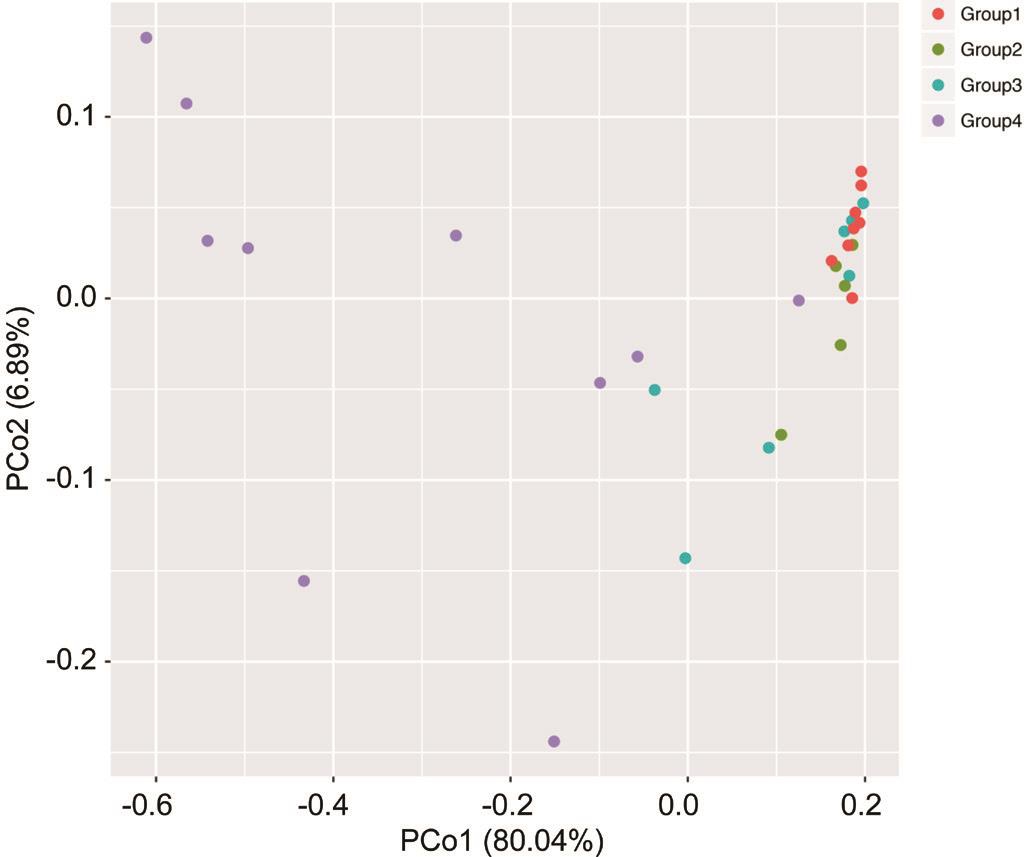
Beta Diversity Analysis Beta diversity refers to species diversity among different environmental groups. Beta diversity and alpha diversity together constitute the biological heterogeneity of overall diversity or a certain environmental community. PCoA represents the phylogenetic distance between samples. The beta diversities in different groups were calculated and visualized through PCoA analyses using the weighted UniFrac distances of 16S rDNA gene between microbial communities in the four groups (Figure 3).Compared with the other three groups, significant difference was observed in Group 4.
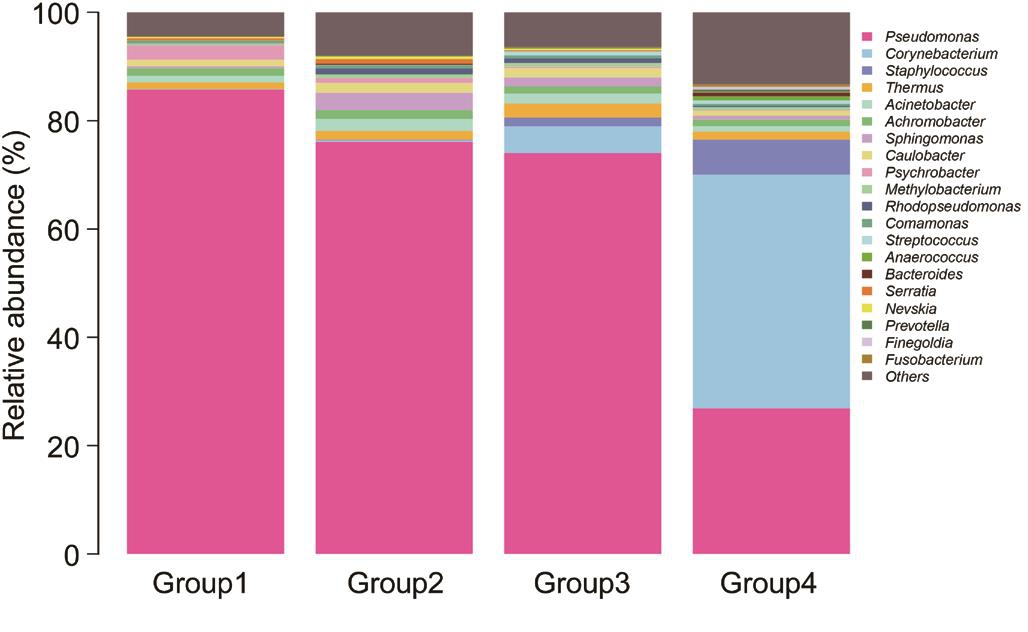
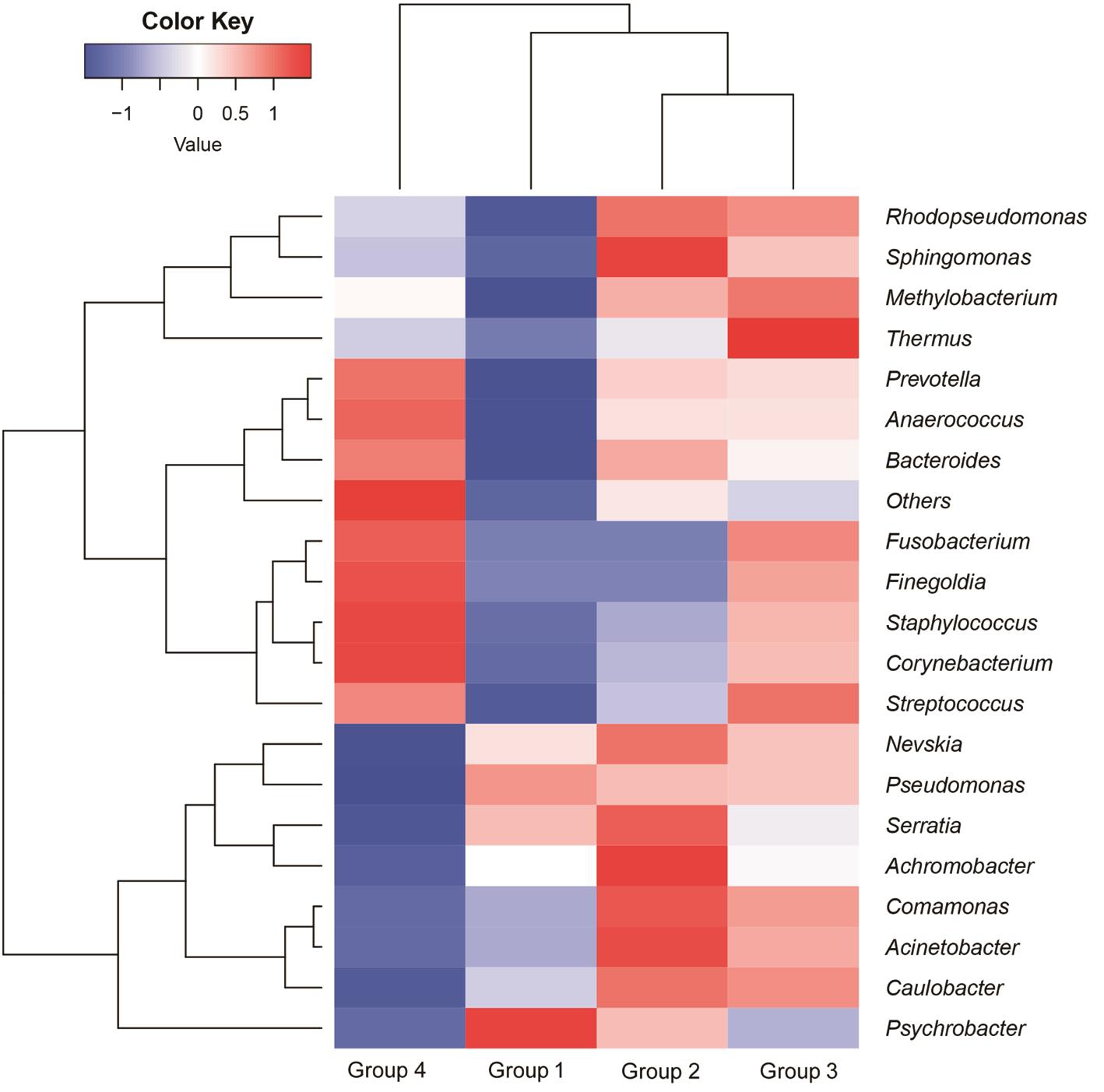
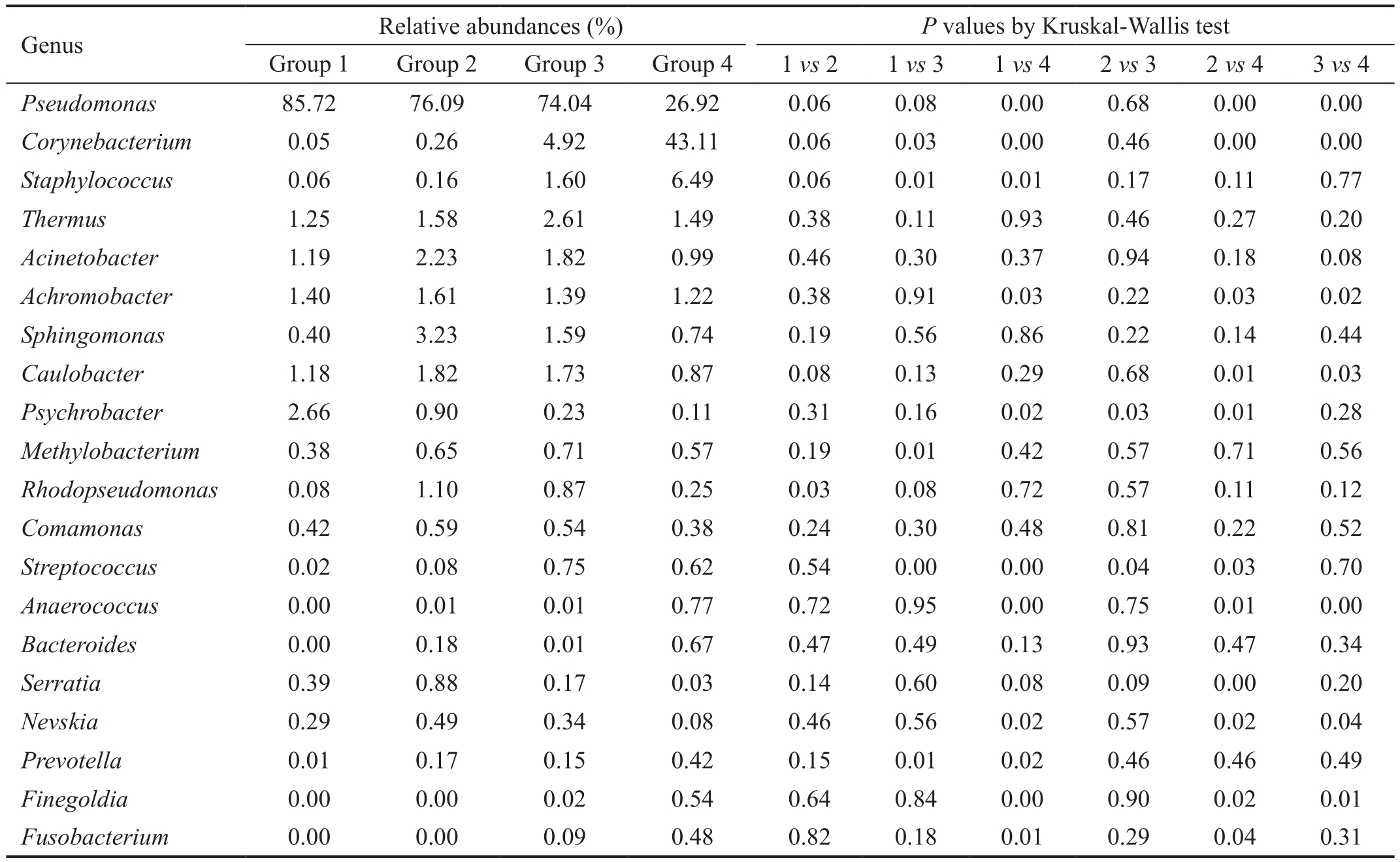
Dominant Genus Analysis At a 97% confidence threshold in the RDP classifier, the 16S rDNA gene sequencing reads were classified into 16 bacterial phyla, 111 families, and 245 genera in all samples, with 67.75±7.79 (range, 54-79)genera detected for each individual in Group 1, 73.80±13.44(range, 55-91) genera in Group 2, 74.57±14.14 (range, 52-93)genera in Group 3, and 89.60±27.49 (range, 44-131) genera in Group 4. The dominant genera were analyzed in Group 1,2, 3 and 4, respectively. As shown in Table 1 and Figure 4,for the healthy eyes (Group 4), the dominant genera (≥1%)included Corynebacterium (43.11%), Pseudomonas (26.92%),Staphylococcus (6.49%), Thermos (1.49%), Achromobacter(1.22%) and Acinetobacter (0.99%). For the fellow eyes(Group 3), the dominant genera (>1%) involved Pseudomonas(74.04%), Corynebacterium (4.92%), Thermus (2.61%),Acinetobacte (1.82%), Caulobacter (1.73%), Staphylococcus(1.60%), Sphingomonas (1.59%) and Achromobacter(1.39%). On the other hand, for the conjunctival sac from the affected eyes (Group 2), the dominant genera (>1%)included Pseudomonas (76.09%), Sphingomona (3.23%),Acinetobacter (2.23%), Caulobacter (1.82%), Achromobacter(1.61%), Thermus (1.58%) and Rhodopseudomonas (1.10%).Furthermore, for the corneal ulcer from affected eyes (Group 1),the dominant genera (>1%) included Pseudomonas (85.72%),Psychrobacter (2.66%), Achromobacter (1.40%), Thermus(1.25%), Acinetobacter (1.19%), and Caulobacter (1.18%).Taken together, the results suggested the dominant genera with significantly different distribution in different groups.
Bacterial Composition Analysis To assess the microbial community alterations, the relative abundance in genus levels was compared and analyzed among 4 groups (Figure 5).Among these genera, 12 genera with significant difference in different groups were found (P<0.05). In our study, the top 5 genera in the ocular surface of the healthy eyes (Group 4) were Pseudomonas Corynebacterium, Staphylococcus, Thermus and Achromobacter, which account for over 70% of the microbial community. Compared with the healthy eyes (Group 4), the lower abundance of Corynebacterium both in affected and fellow eyes of patients suffering FK (Groups 1, 2 and 3) was observed, respectively (P<0.05). The reduced abundance of Staphylococcus both in affected and fellow eyes of patients suffering FK was also detected, but the significant difference only in Group 1 vs Group 4 (P<0.05). While the abundance of Pseudomonas, Achromobacter, Caulobacter, Psychrobacter,Serratia and Nevskia both in affected and fellow eyes of patients with FK was much higher than that in the healthy eyes, respectively (P<0.05). Collectively, the patients suffering FK displayed disrupted conjunctival microbiome, and these alterations may be implicated in the pathogenesis of FK.
Table 1 Relative abundances of the top 20 Genera (mean) and the pairwise comparison
?
DISCUSSION
Diverse microbial communities are colonized on the human ocular surface like other body sites. Ocular microbiome plays a part not only in maintaining homeostasis of ocular surface, but also in the pathogenesis of ocular diseases. However, whether ocular microbiome is relevant to ocular infectious diseases remains largely unknown. In this baseline report, we delineated the differences in the ocular microbiome of the patients with FK in relation to those with healthy eyes. Both the affected and contralateral eyes in the patients with FK showed a decreased bacterial diversity and altered composition, with lower abundances of Corynebacterium and Staphylococcus and higher abundances of Pseudomonas and Psychrobacter.
Our previous investigation identified Corynebacterium,Pseudomonas, Staphylococcus, Acinetobacter and Streptococcus as the “core genera” in the normal conjunctival microbiota[14].In the current study, however, we found that the eyes with FK showed lower abundances of Corynebacterium (0.26%vs 43.11%) and Staphylococcus (0.16% vs 6.49%) compared with the normal eyes. Significantly decreased abundances of Corynebacterium (4.92% vs 43.11%) and Staphylococcus(1.60% vs 6.49%) were also observed in the fellow eyes of the patients with FK compared with the healthy eyes, which predict a higher susceptibility to fungal infection. Corynebacterium mastiditis and Staphylococcus epidermidis were reported to be skin commensal[9,17] and could afford heterologous protection against pathogenic fungal infections through induction of IL-17+γδ+ T and IL-17A+CD8+ T cells, respectively. St Leger et al[12] found that Corynebacterium mastiditis colonized the mouse conjunctiva and protected it against corneal infection by triggering γδ+ T cell-dependent IL-17 response. Our present study provided compelling clinical evidence that the decreased abundances of Corynebacterium and Staphylococcus on the ocular surface contributed to the pathogenesis of FK. Further studies are needed to determine the roles of microbiota in the establishment and maintenance of ocular immune homeostasis and to investigate what triggers the changes of ocular surface bacterial structure in the contralateral eyes.
Our data also disclosed that the relative proportions of some genera were higher in patients with FK than in healthy persons,such as Pseudomonas (76.09% vs 26.92%), Achromobacter(1.61% vs 1.22%), Caulobacter (1.82% vs 0.87%), and Psychrobacter(0.90% vs 0.11%) (P<0.05). Pseudomonas was frequently identified as bacterial pathogens on the human ocular surface,and involved in secondary infection of FK[18]. But some studies also indicated that Pseudomonas has an anti-fungal activity through its secondary metabolites[19-20]. However, it is still unclear what roles the increased Pseudomonas plays in the pathogenesis of FK, which needs to be further investigated.Caulobacter can be recovered from many freshwater facilities,but they are not a part of the normal microbial fl ora of the skin and have rarely been found to be pathogenic in humans. The special requirements for growth, the low temperatures, and the NaCl intolerance suggest that several species of Caulobacter may be overlooked in the clinical microbiology laboratory[21].Similarly, Achromobacter was found in contact lens users,but ocular infections due to Achromobacter are extremely uncommon[22]. Moreover, the mean relative abundance of Psychrobacter in different groups was 2.66%, 0.90%, 0.23%,and 0.11%, respectively, suggesting an increased abundance on the ocular surface with fungal infection. Psychrobacter was found in the extreme environment, with the potential as a resource of enzymes which were able to maintain its activity and stability at low temperature[23]. It was reported to display an anti-fungal activity through their metabolites such as important enzymes (cellulase, protease, lipase, and amylase)[24], and be applied to the control of emerging fungal diseases in wild and farmedfishes[25]. All of the results suggested Pseudomonas and Psychrobacter may be a source of novel antibiotics that can find medicinal use in antifungal therapy for FK in the future.
The limitations of the present study must be acknowledged.Fifirst, although our results were supported statistically, the number of subjects was insufficient. Our team is undertaking further ocular microbiome analyses with more biological samples. Second, 16S rDNA sequencing method we used in this study prevented resolution at the species level, which is important in identifying the bacterial composition. More studies at the species or strain levels will be required to test the validity of our results. Third, although we observed the decreased bacterial diversity and altered bacterial composition in the conjunctiva of eyes with FK, further animal experiments and in vitro investigations are needed to uncover the associations of altered bacteria, such as Corynebacterium and Staphylococcus, with FK and to elucidate the mechanism underlying them. Moreover, analyses of the mycobiome, which is closely related to health and disease analysis[26-27], in the eye through internal transcribed spacer sequencing also necessary for a full undefirstanding of the pathogenesis of FK.
To our knowledge, this is the fifirst study to investigate the conjunctival bacterial communities in FK using 16S rDNA PCR-based gene sequencing metagenomics analysis. The finding of decreased bacterial diversity and altered microbial community of the ocular conjunctiva in patients with FK provides novel clues for the pathogenesis of FK.
ACKNOWLEDGEMENTS
Foundations: Supported by the National Natural Science Foundation of China (No.81670839); the Key Research and Development Plan of Shandong Province (No.2018CXGC1205).
Conflicts of Interest: Ge C, None; Wei C, None; Yang BX,None; Cheng J, None; Huang YS, None.
REFERENCES
1 Dan J, Zhou QJ, Zhai HL, Cheng J, Wan L, Ge C, Xie LX. Clinical analysis of fungal keratitis in patients with and without diabetes. PLoS One 2018;13(5):e0196741.
2 Wen XF, Miao L, Deng YH, Bible PW, Hu X, Zou YL, Liu Y, Guo SX,Liang JR, Chen TT, Peng GH, Chen W, Liang LY, Wei L. The influence of age and sex on ocular surface microbiota in healthy adults. Invest Ophthalmol Vis Sci 2017;58(14):6030-6037.
3 Kugadas A, Christiansen SH, Sankaranarayanan S, Surana NK,Gauguet S, Kunz R, Fichorova R, Vorup-Jensen T, Gadjeva M. Impact of microbiota on resistance to ocular pseudomonas aeruginosa-induced keratitis. PLoS Pathog 2016;12(9):e1005855.
4 Kugadas A, Gadjeva M. Impact of microbiome on ocular health. Ocul Surf 2016;14(3):342-349.
5 Kugadas A, Wright Q, Geddes-McAlister J, Gadjeva M. Role of microbiota in strengthening ocular mucosal barrier function through secretory IgA. Invest Ophthalmol Vis Sci 2017;58(11):4593-4600.
6 de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S,Streckfus CF, Hutchinson DS, Ajami NJ, Petrosino JF, Pflugfelder SC.Altered mucosal microbiome diversity and disease severity in sjögren syndrome. Sci Rep 2016;6:23561.
7 Lee SH, Oh DH, Jung JY, Kim JC, Jeon CO. Comparative ocular microbial communities in humans with and without blepharitis. Invest Ophthalmol Vis Sci 2012;53(9):5585-5593.
8 Zhang HK, Zhao FX, Hutchinson DS, Sun WF, Ajami NJ, Lai SJ, Wong MC, Petrosino JF, Fang JH, Jiang J, Chen W, Reinach PS, Qu J, Zeng CQ,Zhang DK, Zhou XT. Conjunctival microbiome changes associated with soft contact lens and orthokeratology lens wearing. Invest Ophthalmol Vis Sci 2017;58(1):128.
9 Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C,Conlan S, Himmelfarb S, Byrd AL, Deming C, Quinones M, Brenchley JM, Kong HH, Tussiwand R, Murphy KM, Merad M, Segre JA, Belkaid Y. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015;520(7545):104-108.
10 Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan XW,Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 2015;21(7):808-814.
11 Tanure MA, Cohen EJ, Sudesh S, Rapuano CJ, Laibson PR. Spectrum of fungal keratitis at wills eye hospital, philadelphia, pennsylvania.Cornea 2000;19(3):307-312.
12 St Leger AJ, Desai JV, Drummond RA, Kugadas A, Almaghrabi F, Silver P, Raychaudhuri K, Gadjeva M, Iwakura Y, Lionakis MS,Caspi RR. An ocular commensal protects against corneal infection by driving an interleukin-17 response from mucosal γδ T cells. Immunity 2017;47(1):148-158.e5.
13 Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM.Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 1998;5(10):R245-R249.
14 Huang Y, Yang B, Li W. Defining the normal core microbiome of conjunctival microbial communities. Clin Microbiol Infect 2016;22(7):643.e7-643643.e12.
15 Yang BX, Li H, Kong FF, Huang YS. Identification of pathogenic bacteria in aqueous and vitreous of endophthalmitis by 16S rDNA sequencing technique. Chin J Exp Ophthalmol 2016;34(10):883-887.
16 Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27(21):2957-2963.
17 Dong QF, Brulc JM, Iovieno A, Bates B, Garoutte A, Miller D,Revanna KV, Gao X, Antonopoulos DA, Slepak VZ, Shestopalov VI.Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci 2011;52(8):5408.
18 Krishnan T, Sengupta S, Reddy PR, Ravindran RD. Secondary pseudomonas infection of fungal keratitis following use of contaminated natamycin eye drops: a case series. Eye (Lond) 2009;23(2):477-479.
19 Ligon JM, Hill DS, Hammer PE, Torkewitz NR, Hofmann D,Kempf HJ, van Pée KH. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag Sci 2000;56(8):688-695.
20 Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R,Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix MA, Schulenburg H.The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol 2016;14:38.
21 Justesen US, Holt HM, Thiesson HC, Blom J, Nielsen XC, Dargis R,Kemp M, Christensen JJ. Report of the first human case of caulobacter sp.infection. Journal of Clinical Microbiology 2007;45(4):1366-1369.
22 Almenara Michelena C, del Buey MÁ, Ascaso FJ, Cristóbal JÁ.Keratitis due to achromobacter xylosoxidans in a contact lens user. Eye &Contact Lens: Science & Clinical Practice 2017:1.
23 Loperena L, Soria V, Varela H, Lupo S, Bergalli A, Mairan GG,Pellegrino A, Bernardo A, Calviño A, Rivas F, Batista S. Extracellular enzymes produced by microorganisms isolated from maritime Antarctica.World J Microbiol Biotechnol 2012;28(5):2249-2256.
24 Bibi F, Ullah I, Alvi SA, Bakhsh SA, Yasir M, Al-Ghamdi AAK,Azhar EI. Isolation, diversity, and biotechnological potential of rhizo- and endophytic bacteria associated with mangrove plants from Saudi Arabia.Genet Mol Res 2017;16(2).
25 Lowrey L, Woodhams DC, Tacchi L, Salinas I. Topographical mapping of the rainbow trout (oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol 2015;81(19):6915-6925.
26 Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA,Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013;498(7454):367-370.
27 Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012;336(6086):1314-1317.
Correspondence to: Yu-Sen Huang. Shandong Eye Institute, 5 Yanerdao Road, Qingdao 266071, China. Huang_yusen@126.com Received: 2018-09-12 Accepted: 2018-11-07
● KEYWORDS: conjunctiva microbiota; bacteria; metagenomics;16S rDNA; fungal keratitis
DOl:10.18240/ijo.2019.02.02
Citation: Ge C, Wei C, Yang BX, Cheng J, Huang YS. Conjunctival microbiome changes associated with fungal keratitis: metagenomic analysis. Int J Ophthalmol 2019;12(2):194-200