INTRODUCTION
M arfan syndrome (MFS; MIM #154700), incidence 1/5000-1/10 000, is an autosomal dominant disorder in which fibrous connective tissue all over the body can be affected[1-2]. Mostly presented manifestations are in cardiovascular, skeletal, ocular, pulmonary and nervous systems. Ocular signs include ectopia lentis, high myopia,peripheral retinal degeneration and rhegmatogenous detachment,etc[3-5]. Diagnosis of MFS is based on both clinical criteria and genetic test[6-7]. For example, a major clinical manifestation plus an FBN1 mutation is enough to confirm the diagnosis[8].FBN1 (location: 15q-21, 65 exons) is the major causative gene accounting for 90% of MFS cases[9-10]. FBN1 codifies fibrillin-1,a 350-kDa glycoprotein macromolecule that polymerizes to form microfibrils, which serves mechanical support in connective tissue throughout the body[11-12]. It also regulates microfibril assemble and stability[12]. Therefore, FBN1 mutations leads to fibrillin-1 disruption, microfibril malformation, and eventually the attenuation of connective tissues[13-14].Most FBN1 mutations are unique for specific MFS families,while 15% of them recur among different pedigrees[11]. We report a missense mutation in exon 32 of FBN1 (c.3932A>G),resulting in a tyrosine to cysteine change at codon 1443(p.Y1311C). This mutation was identified in a 4-generation Chinese family with typical MFS signs together with a peculiar macular degeneration. To the best of our knowledge, neither the mutation nor this macular lesion has been reported in MFS before.
SUBJECTS AND METHODS
Ethical Approval This study was approved by the Ethics Committee on Clinical Investigations of the Second Xiangya Hospital of Central South University, and carried out in accordance with the Declaration of Helsinki for Human Subjects. All participants were given informed consent, and then underwent ophthalmologic, cardiovascular and systemic physical examinations.
Table 1 Clinic features of the patients in the pedigree
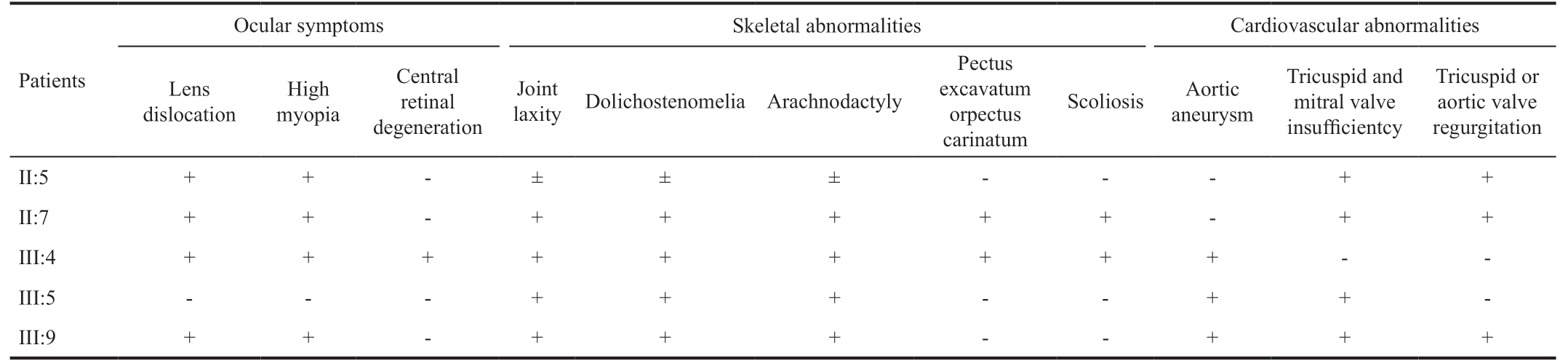
Ocular symptoms Skeletal abnormalities Cardiovascular abnormalities Patients Lens dislocation Joint laxity Dolichostenomelia Arachnodactyly High myopia Central retinal degeneration Pectus excavatum orpectus carinatum Scoliosis Aortic aneurysm Tricuspid and mitral valve insufficientcy Tricuspid or aortic valve regurgitation II:5 ++-± ± ± ---++II:7 + + - + + + + + - + +III:4 + + + + + + + + + - -III:5 - - - + + + - - + + -III:9 + + - + + + - - + + +
Patients and the Clinical Data Diagnosis of MFS were confirmed according to the revised Ghent criteria[4]. Clinical data were collected from 14 living family members (five patients: II:5, II:7, III:4, III:5, III:9; nine unaffected family members: II:1, II:6, II:8, III:1, III:3, III:10, III:11, IV:1 and IV:2) in the pedigree, all family members received systemic review and ophthalmic examination for MFSrelevant abnormalities, including physical examination, chest X-ray, cardiac ultrasonography, best corrected visual acuity(BCVA), slit lamp examination and indirect ophthalmoscopy.Photography of the anterior segment and fundoscopy were recorded if any abnormality presented. Optical coherence tomography (OCT) images (Topcon 3D 2000, Japan) centered on the fovea showed structural changes.
DNA Sample Collection and Sanger Sequencing of FBN1 Gene Genomic DNA extraction were performed using peripheral blood lymphocytes following the standard phenolchloroform method. DNA samples of patients were used for polymerase chain reaction (PCR) amplification of exon 24,25, 26, 27, 28, 30, 31, and 32. DNA samples of other family members and 100 normal controls were amplified on exon 32.Primer sequences as follow:
FBN1-24-F: 5’-GGGCTCGTTCTGGTTGCTA-3’, FBN1-24-R: 5’-TGCTGCTTACTATTTGAAAGACTGT-3’, FBN1-25-F: 5’-TTATTAGGCAAGGATACTTACCC-3’, FBN1-25-R: 5’-GGACTTCTTGGACCAAACAGT-3’, FBN1-26-27F: 5’-AAGGCTAGAAATGTTTACAAAGTCA-3’, FBN1-26-27R: 5’-TCCCTCATTCTTTCTACCTCAGT-3’, FBN1-28-29-F: 5’-GCATTTTGGTTTTAGTCTGAT-3’, FBN1-28-29-R: 5’-CAGGGAAACAAGAAAGATAAG-3’, FBN1-30-F: 5’-ATCCCACCATGAGGGTAGAG-3’, FBN1-30-R: 5’-TATGCAGGCAATTTGAACTTC-3’, FBN1-31-F: 5’-TTTACCAAGGATAACCCAATG-3’, FBN1-31-R: 5’-AGTCAAAGCAGAAGGAGGGT-3’, FBN1-32-F:5’-ATGAAAGCCAGTCTGAATAATG-3’, FBN1-32-R:5’-TTCAAAGAAGTGGAAGCTAAAT-3’.
PCR was performed in a 50 µL reaction mixture containing 5 µL Pfu DNA Polymerase (Sangon Biotech Co., Ltd., Shanghai,China, SC0014), 5 µL 10×PCR Buffer without Mg2+, 20 pmol of each primer, 100 ng of genomic DNA, 5 µL MgCl2(25 mmol/L), 1 µL 10 mmol/L dNTP. The amplification conditions consisted of denaturation at 95℃ for 3min, followed by 35 cycles of denaturation at 94℃ for 30s, annealing at 55℃-60℃for 35s, and extension at 72℃ for 40-50s. A final extension was stranded at 72℃ for 7min. The PCR products were then purified by DNA purification kit (Sangon Biotech Co., Ltd.,Shanghai, China, SK1141). Sanger sequencing was performed on Applied Biosystems 3730 DNA Analyzers (Hudson, New Hampshire, USA). Sequencing results were analyzed by DNASTAR Lasergene software (Madison, Wisconsin, USA).
Mutant Analysis Analysis of the conservatism of nonsynonymous variant was performed by multiple sequence alignment tools Clustal Omega (http://www.ebi.ac.uk/Tools/msa/). Software SIFT (http://sift.jcvi.org/www/SIFT_enst_submit.html) were used to perform functional prediction,PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and Mutation Taster (http://www.mutationtaster.org/) to score nonsynonymous variant.
RESULTS
Clinical Findings The proband is from a 4-generation consanguineous Chinese family consisting of 13 men and 8 women (pedigree shown in Figure 1A). Of 5 individuals in this pedigree were confirmed with the diagnosis MFS, clinical features were shown in Table 1. The median onset age of the patients was 29y, ranging from 22 to 62y. All 5 patients had typical ocular, skeletal and cardiovascular manifestations.Common MFS skeletal abnormalities like dolichostenomelia,pectus excavatum/carinatum, and scoliosis, joint laxity and arachnodactyly were observed in all affected family members.Cardiovascular findings including aortic aneurysm, tricuspid/mitral valve insufficiency, tricuspid/aortic valve regurgitation,mitral valve prolapse were noted in patient III:5 and III:9, both received Bentall surgery. Shared ophthalmic findings include high myopia, bilateral lens dislocation, and peripheral retinal degeneration. The proband III:4 presents with an acute angleclosure glaucoma secondary to anteriorly dislocated lens after mild trauma in the left eye (Figure 1B-1D). She had lensectomy and scleral fixated intraocular lens (SF-IOL) implantation in both eyes. Her visual acuity is 0.4 for her better eye, and 0.2 for worse eye. Moreover, the proband and her brother (III:5)present an unusual bilateral macular degeneration appearance in both eyes (Figure 1E-1L). Patient II:3 deceased of dissecting aneurysm at the age of 30, his off-springs III:6, III:7, and III:8 all died in utero or at very early age with “reasons unidentified”.
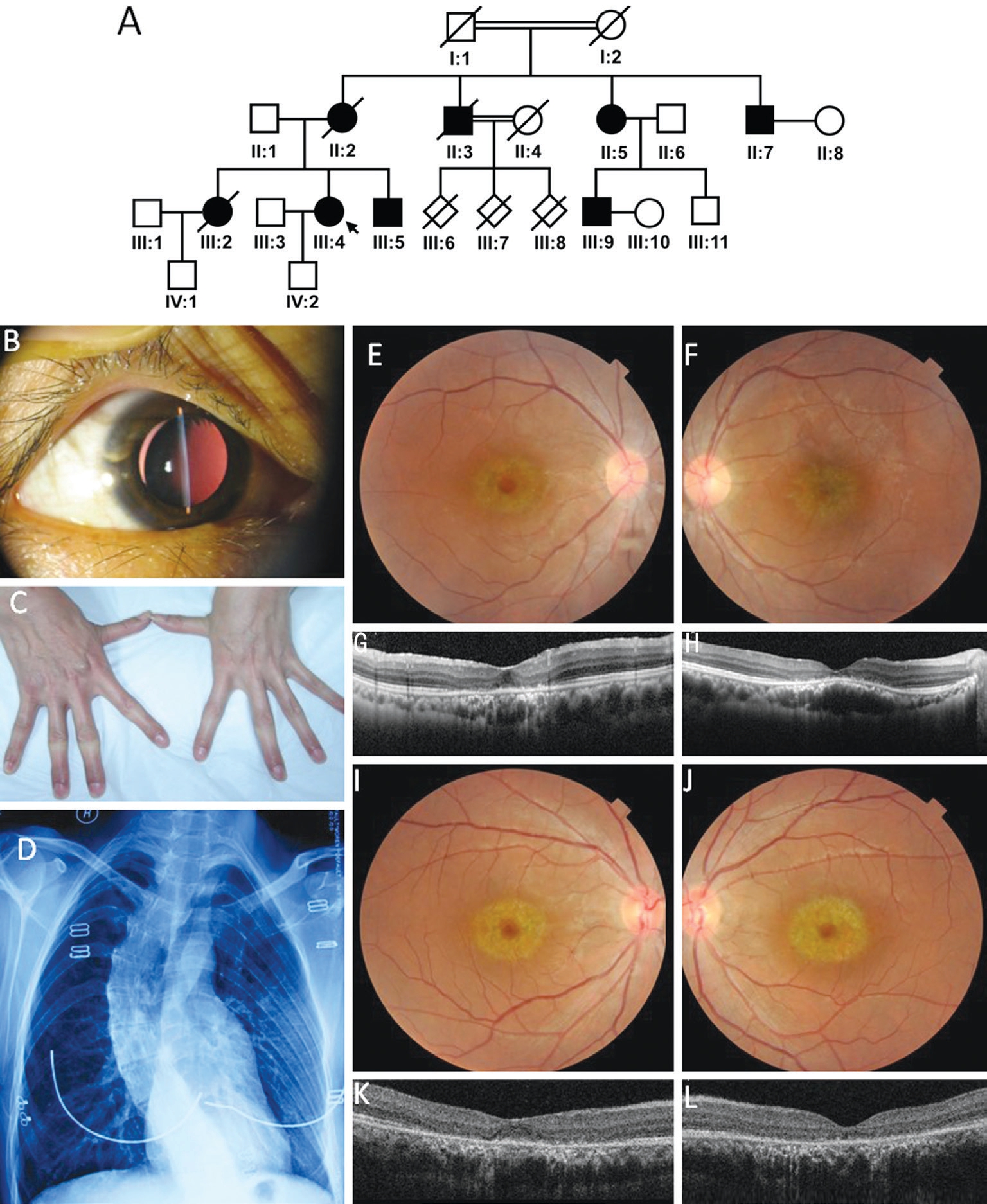
Figure 1 Pedigree and phenotypes of the MFS family A: Pedigree of the family; B: Ectopia lentis in the proband; C: Arachnodactyly and the laxity of the joints; D: Scoliosis shown on chest X-ray; E-L: Unusual bilateral macular degeneration of the proband and her brother; E, F:Fundus photography showed symmetrical macular degeneration of right eye (E) and left (F) of the proband; G, H: OCT scan through the lesion showed marked thinning of the outer retinal layers (attenuation and disruption of the photoreceptor and retinal pigment epithelium layer). Also note a subretinal accumulation of hyperreflective material under the fovea. Subfoveal choroid vessels seems to have enlarged lumen in proband’s left eye (H). Outside this area, retinal structures appear intact; I-L: Similar changes were found in both eyes of the proband’s brother (III:5); I, J:Fundus photography showed symmetrical macular degeneration of both eye and similar to the proband; K, L: OCT showed similar but milder thinning of the outer retinal layers, disruption of the photoreceptor and accumulation of hyperreflective material between the neurosensory retina and RPE in the fovea area.
Genetic Analysis Identified FBN1 Heterozygous Missense Mutation DNA sample of our proband III:4 were performed PCR and Sanger sequencing to find out mutations in exons 24,25, 26, 27, 28, 30, 31, and 32, for mutants in these exons are often associated with early onset, prominent MFS phenotypes.Genetic analysis detected a heterozygous missense mutation c.3932A>G (hg37, NM_000138) in our proband, which lead to a mutant protein p.Y1311C. The mutation has not been reported in dbSNP database (https://www.ncbi.nlm.nih.gov/snp) and 1000 Genome (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/). We performed Sanger sequencing to confirm the co-segregation status in all patients and normal relatives in the family. We found the mutation was consistent with the phenotypes in the family. We assessed this variant in 100 normal controls furtherly and found no mutation at this locus (Figure 2A).

Figure 2 Identification of the c.3932A>G mutation in FBN1 on exon 34 A: Presentative sequencing results of the mutation in all affected members and the corresponding normal sequence in the unaffected family members; B: Evolutionary conservation analysis for p.Y1311C residue. Asterisk: Complete identical amino acid; Codon: Highly conserved amino acid; Dot: Moderate conserved amino acid. Red line marked mutated amino acid residues.
Mutation Analysis of p.Y1311C To explore possible impact of the missense mutation on FBN1 protein function, we performed the conservation and function prediction through Clustal Omega,MutatinTaster, Polyphen-2 and SIFT on our mutant. Clustal Omega result predicted asterisks in the amino acid position 1311 (Figure 2B), which indicated highly conserved amino acid on this site. As shown in Table 2, MutationTaster predicted that p.Y1311C was disease causing nucleotide substitution. Both Polyphen-2 and SIFT results demonstrated that the mutation was very likely a damaging nucleotide variation. Conservative functional analysis tools predicted that p.Y1311C mutation probably affected the structure and function of human fibrillin-1 protein.
DISCUSSION
Over 3000 FBN1 mutations have been reported in the Universal Marfan Mutation Database (UMD-FBN1; http:/www.umd.be/FBN1/), in which 60.3% were missense mutations[15]. In this study we have identified a novel FBN1 heterozygous missense mutation (c.3932A>G, p.Y1311C) in our pedigree. This mutation causes tyrosine to cysteine substitution at the 1311th amino acid, which could results in abnormality of fibrillin-1 in the heterozygous affected individuals. Fibrillin-1 molecular structure comprises of 47 epidermal growth factor-like (EGF)domains and seven transforming growth factor-β1 binding protein-like (TB) domains, most mutations of FBN1 appeared in the EGF domains[16] which could disrupt formation of microfibril, the vital component of extracellular matrix (ECM)and collagen universally expressed in connective tissues in major vessels, cartilages, tendons, corneas, zonules, etc[17-18].Fibrillin-1 also functions as an important regulator of TGFβ pathway through forming complex with TGFβ, latent TGFβ binding protein (LTBPs), and microfibrils[19-21]. Since our p.Y1311C mutation was predicted to disrupt the biological function or structure of FBN1 protein, it is very likely to disrupt microfibril formation and subsequently cause MFS.Individuals in this pedigree with heterozygous mutation have MFS phenotypes, which is coincident with other study that missense mutation often cause ectopia lentis[4,6]. Moreover, our pedigree showed a very unusual phenotype of symmetrical bilateral macular degeneration in addition to common phenotype like ectopia lentis, arachnodactyly and scoliosis.Fundus photography showed symmetrical bilateral macular lesions in the proband and her living brother, OCT further revealed an uncommon lesion for MFS in the outer retinal layers: thinning of outer nuclear layer, disruption of external limiting membrane and photoreceptor layers, and accumulation of hyper-reflective material between the neurosensory retina and RPE under the fovea. These changes all imply degenerated photoreceptors in the foveal area, hyperreflective material may correspond to clumping of abnormal photoreceptor outer segments material. As far as we are concerned, this feature has never been reported in Marfan syndrome. Our hypothesis is the microfibrils in photoreceptor-RPE interdigitation and Bruch’s membrane is also affected in this mutation, so the RPE-Bruch’s membrane-choriocapillaris complex is unhealthy and degenerates with time.
Table 2 Conservative and functional analysis on c.3932A>G (p.Y1311C) missense mutation in FBN1
P: Probably damaging; D: Damaging.
Mutant Exon No. MutationTaster PolyPhen-2 SIFT c.3932A>G 32 Disease causing 0.999, P 0.000, D
In our study, a novel mutation of FBN1 c.3932A>G in exon 32 was identified in a Chinese consanguineous MFS family with an unusual phenotype of macular degeneration. The result not only expands our knowledge of FBN1 mutations, but also is helpful in definite diagnosis of uncertain MFS cases. The genotype-phenotype relationship of the new mutation and its unusual sign of bilateral macular degeneration needs to be identified by further study.
ACKNOWLEDGEMENTS
We are grateful to all the patients, their unaffected family members, and the healthy volunteers for their participation.The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions: Li Y and Zhao Y analyzed and interpreted the patient data regarding the Marfan syndrome.Ouyang PB, Cao J and Zhao Y performed the patients and the clinical data. Zhang LS performed the genetic analysis.Peng YQ and Cao J was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Foundations: Supported by National Natural Science Foundation of China (No.81300798); Project supported by the Natural Science Foundation of Hunan Province, China(No.2018JJ3737); Department of Science and Technology,Hunan (No.2015TP2007).
Conflicts of Interest: Ouyang PB, None; Zhao Y, None;Peng YQ, None; Zhang LS, None; Cao J, None; Li Y, None.
1 Judge DP, Dietz HC. Marfan’s syndrome. Lancet 2005;366(9501):1965-1976.
2 Dietz H: Marfan syndrome. 2017.In: M Adam, HH Ardinger, RA Pagon, SE Wallace, LJH Bean, K Stephens, A Amemiya editors. GeneReviews.2017.Available at: http://www.ncbi.nlm.nih.gov/books/NBK1335/.
3 Maumenee IH. The eye in the Marfan syndrome. Trans Am Ophthalmol Soc 1981;79:684-733.
4 Loeys BL, Dietz HC, Braverman AC, Callewaert BL, de Backer J,Devereux RB, Hilhorst-Hofstee Y, Jondeau G, Faivre L, Milewicz DM,Pyeritz RE, Sponseller PD, Wordsworth P, De Paepe AM. The revised Ghent nosology for the Marfan syndrome. J Med Genet 2010;47(7):476-485.
5 Dean JC, Loeys B. Marfan syndrome and related disorders. In:Cardiovascular Genetics and Genomics. Springer 2018:pp589-615.
6 Zhurayev R, Proost D, Zerbino D, Fedorenko V, Meester JAN, van Laer L, Loeys BL. Identification of FBN1 gene mutations in Ukrainian Marfan syndrome patients. Genet Res 2016;98:e13.
7 Groth KA, Hove H, Kyhl K, Folkestad L, Gaustadnes M, Vejlstrup N,Stochholm K, Østergaard JR, Andersen NH, Gravholt CH. Prevalence,
incidence, and age at diagnosis in Marfan Syndrome. Orphanet J Rare Dis 2015;10:153.
8 Pepe G, Giusti B, Sticchi E, Abbate R, Gensini GF, Nistri S. Marfan syndrome: current perspectives. Appl Clin Genet 2016;9:55-65.
9 Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr Opin Genet Dev 2007;17(3):252-258.
10 Baetens M, van Laer L, de Leeneer K, Hellemans J, de Schrijver J, van de Voorde H, Renard M, Dietz H, Lacro RV, Menten B, van Criekinge W,de Backer J, de Paepe A, Loeys B, Coucke PJ. Applying massive parallel sequencing to molecular diagnosis of Marfan and Loeys-Dietz syndromes.Hum Mutat 2011;32(9):1053-1062.
11 Loeys B, De Backer J, Van Acker P, Wettinck K, Pals G, Nuytinck L,Coucke P, De Paepe A. Comprehensive molecular screening of the FBN1 gene favors locus homogeneity of classical Marfan syndrome. Hum Mutat 2004;24(2):140-146.
12 Abdelrahim M, Khalid KE, Faris ME, et al. Review of FBN1 gene role in Marfan syndrome presentations insilico analysis. American Journal of Biomedical Research 2016;4(1):5-12.
13 Zeyer KA, Reinhardt DP. Engineered mutations in fibrillin-1 leading to Marfan syndrome act at the protein, cellular and organismal levels. Mutat Res Rev Mutat Res 2015;765:7-18.
14 Aragon-Martin JA, Child AH. Marfan Syndrome (MFS): Inherited Microfibrillar Disorder Caused by Mutations in the Fibrillin-1 Gene. In: Diagnosis and Management of Marfan Syndrome. Springer 2016:pp233-243.
15 Wang F, Li B, Lan L, Li L. C596G mutation in FBN1 causes Marfan syndrome with exotropia in a Chinese family. Mol Vis 2015;21:194-200.
16 Xiao Y, Liu X, Guo X, Liu L, Jiang L, Wang Q, Gong B. A novel FBN1 mutation causes autosomal dominant Marfan syndrome. Mol Med Rep 2017;16(5):7321-7328.
17 Hubmacher D, Tiedemann K, Reinhardt DP. Fibrillins: from biogenesis of microfibrils to signaling functions. Curr Top Dev Biol 2006;75:93-123.
18 Kielty CM, Sherratt MJ, Marson A, Baldock C. Fibrillin microfibrils.In: Advances in protein chemistry. Vol 70. Elsevier 2005:pp405-436.
19 Ramirez F, Sakai LY. Biogenesis and function of fibrillin assemblies.Cell Tissue Res 2010;339(1):71-82.
20 Benke K, Ágg B, Szilveszter B, Tarr F, Nagy ZB, Pólos M, Daróczi L, Merkely B, Szabolcs Z. The role of transforming growth factor-beta in Marfan syndrome. Cardiol J 2013;20(3):227-234.
21 Todorovic V, Jurukovski V, Chen Y, Fontana L, Dabovic B, Rifkin DB.Latent TGF-β binding proteins. Int J Biochem Cell Biol 2005;37(1):38-41.