INTRODUCTION
H ypoxia-induced retinal neovascularization is a major complication of many ocular diseases including diabetic retinopathy, retinopathy of prematurity and retinal vein occlusion, and it universally leads to severe visual loss[1-3]. Numerous studies demonstrated that vascular endothelial growth factor (VEGF) acts as a critical factor in intraocular neovascular diseases, and anti-VEGF agents can effectively suppress intraocular neovascularization in clinical applications[4-7]. Nevertheless, some of the patients were not responding to anti-VEGF therapy, and various drugrelated adverse effects have been reported, including retinal detachment, increased intraocular pressure and macular hole formation[4]. In addition, studies suggest that there are several molecules other than VEGF that contribute to the pathogenesis of retinal neovascularization[8-9]. Oxygen-induced retinopathy(OIR) in mice is a commonly used animal model to investigate the molecule mechanisms of retinal neovascularization[10].Several studies have explored gene expression profiles in mice with OIR, suggesting that a number of genes are involved in the induction of retinal neovascularization[11-13].Moreover, OIR mouse model was also used to explore the roles of macrophages, periostin and interleukins in retinal neovascularization[14-16].
MicroRNAs (miRNAs) are about 22 nucleotides (nt) long and non-coding RNAs that post-transcriptionally regulate coding gene expressions. miRNAs usually binds to the 3’-untranslated region (3’-UTR) of target genes, resulting in either translational inhibition[17] or degradation of mRNAs[18]. miRNAs have been recognized to play crucial regulatory roles in a wide range of physiological and pathological processes[19-21], and are also known to be expressed in a tissue-specific manner[22]. A few studies have found aberrant miRNA expression in different pathological ocular tissues[23-26], suggesting important roles of miRNAs in ocular diseases. Although several similar studies have reported the retinal miRNA expression profile in OIR model[27-29], the mechanisms by which miRNAs regulate target gene expression that induce retinal neovascularization still remain unclear. Thus, we assess miRNA expression profile in retinas of mice with OIR by miRNA microarray and bioinformatically analyze the interaction of target genes of those altered miRNAs in retinal neovascularization.
MATERIALS AND METHODS
Ethical Approval C57BL/6J mice (Hunan SJA Laboratory Animal Co., Ltd, Changsha, Hunan, China) were used in the study. Animal experiments were performed following the Statement on the Use of Animals in Ophthalmic and Vision Research of Association for Research in Vision and Ophthalmology. Institutional Animal Care and Use Committee of Central South University approved the experimental procedures of the study.
Mouse Model of Oxygen-induced Retinopathy and Sample Collection OIR was induced in mice according to previous protocols[10,30-31]. Briefly, pups at postnatal day 7 (P7)accompanied with the nursing mother were exposed to oxygen of 75%, and returned to room air at P12. Pups in room air during the whole period were used as a control group. Mouse retina samples from both groups were collected at P17.
RNA Isolation and miRNA Expression Microarray The total RNA of retinas was isolated using Trizol reagent (Life technologies, NY, USA). Briefly, the retinas from both eyes of a mouse were pooled as a sample, followed by addition of 300 µL of Trizol reagent. After homogenization, the homogenized samples were added with 60 µL of chloroform and were vigorously shaked. Samples were centrifugated at 12 000×g for 15min at 4℃. The aqueous phase was placed into a microtube with 150 µL of 100% isopropanol, followed by centrifugation at 12 000×g for 10min at 4℃. Finally, the resulting pellet containing RNAs was washed with 300 µL of 75% ethanol, and the air-dried RNAs were dissolved in RNasefree water. NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used for measuring the RNA quality and quantity, and denaturing agarose gel electrophoresis was used to determine the RNA integrity.miRNAs labeled by a miRCURY Hy3/Hy5 Power labeling kit(Exiqon, Vedbaek, Denmark) were hybridized by a miRCURY Array (v.19.0, Exiqon), and the array were scanned by an Axon GenePix 4000B microarray scanner (Axon Instruments,Foster City, CA, USA). The raw data were imported into the system of a GenePix Pro 6.0 software (Axon Instruments)for the purpose of grid alignment and data extraction. After normalization, altered miRNAs were identified at fold change≥1.5, and P<0.05. The microarray data were uploaded to the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) for public access (GEO Series accession number GSE115949).
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction Validation for Altered miRNAs The miRNAs were validated by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). Briefly,1.5 µg of total RNA that included the small RNAs was reverse-transcripted using All-in-One™ miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA).qRT-PCR was conducted by using the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA,USA). The expression levels of miR-350-3p, miR-202-3p,miR-711, miR-30c-1-3p, and U6 were measured by qRTPCR using a miRNA qPCR Mix (GeneCopoeia). Primers purchased from GeneCopoeia were defined as follows: miR-350-3p (MmiRQP0982), miR-202-3p (MmiRQP0923), miR-711 (MmiRQP1139), miR-30c-1-3p (MmiRQP0394), and U6(MmiRQP9002). The U6 small nuclear RNA was employed as an endogenous control to normalise the expression levels of miRNAs. The relative expression of miRNAs in mice with OIR was calculated using the median ΔCt value of the normal retina tissues by the 2-ΔΔCt method.
In Silico Analyses We used Targetscan7.1 (http://www.targetscan.org/mmu_71/) and MirdbV5 database (http://mirdb.org/miRDB/) to predict target genes of miRNAs. Those shared target genes between two databases were used for miRNAmRNA network analysis. Gene Ontology (GO) analysis (http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/) were conducted to predict possible biological functions of those target genes of altered miRNAs.
Statistical Analyses The statistical difference of significance was assessed by Student t-test, and P<0.05 was considered as statistically significant throughout the present study.
RESULTS
Altered miRNA Expression in Mice with Oxygen-induced Retinopathy To investigate the difference in retinal miRNAs expression profile between OIR and control mice, we performed the Exiqon microarray with retinas from 6 mice(3 OIRs and 3 controls). miRNAs microarray data analysis suggested that 289 miRNAs were upregulated in OIRs compared to controls with fold change greater than 1.5-fold,and 59 miRNAs were downregulated at more than 1.5-fold(Figure 1A). As showed in Figure 1 and Table 1, among these miRNAs, 3 miRNAs (mmu-miR-350-3p, mmu-miR-202-3p,and mmu-miR-100-5p) were significantly upregulated, and 8 miRNAs (mmu-miR-711, mmu-miR-10a-3p, mmu-miR-201-3p, mmu-miR-383-5p, mmu-let-7c-5p, mmu-let-7e-5p,mmu-miR-30c-1-3p, and mmu-miR-882) were significantly downregulated in mice with OIR. The heatmap revealed distinguishable miRNA expression profile between control and experimental group except for OIR1 (Figure 1B), which may be caused by individual difference. In order to avoid experimental errors, another group of OIR and control mice were used for validation of altered miRNA expression by qRTPCR.
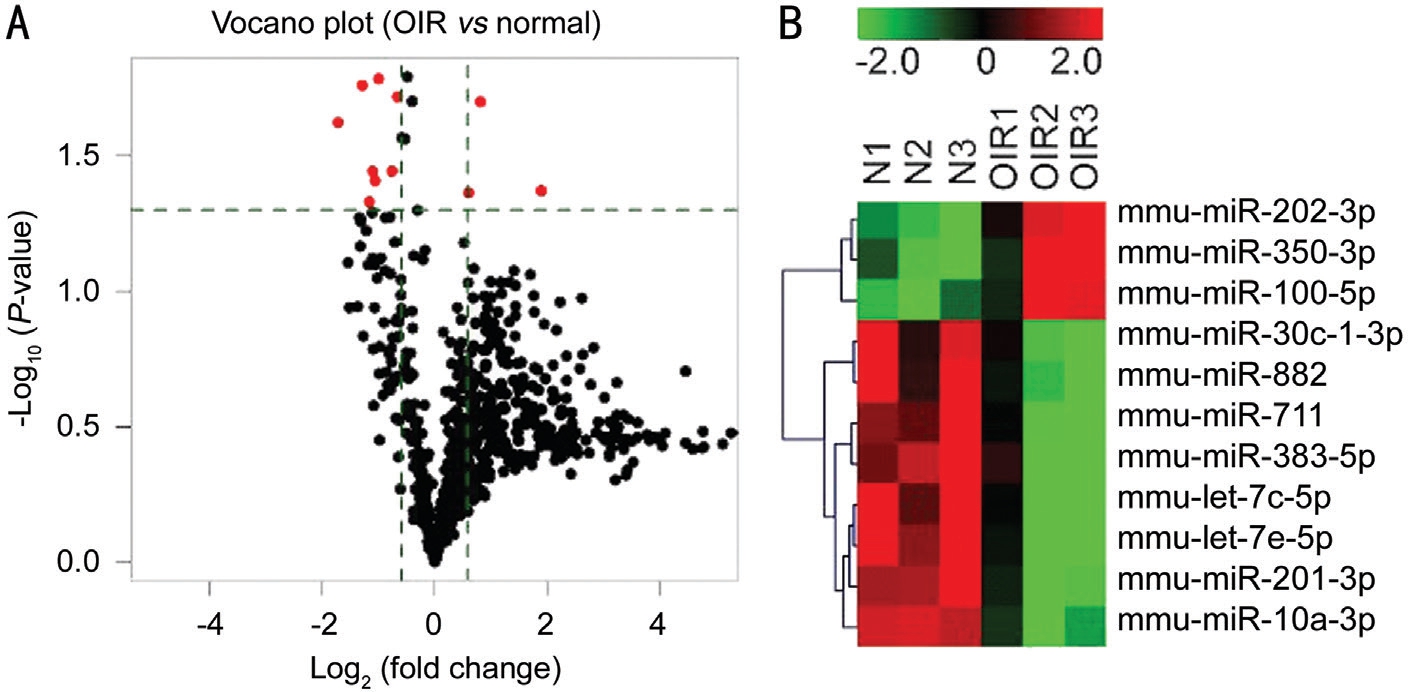
Figure 1 miRNA expression profiles were altered in the mouse retinas with OIR A: The volcano plots illustrates the fold-change values and P-values of altered miRNA expressions in OIR compared with normal retinas. The vertical lines represent to 1.5-fold change of up- and downregulation, the horizontal line represents P=0.05. The red points represent the significantly altered miRNAs; B: The heatmap of altered miRNAs in the OIR and control groups. Red color represents high relative expression, and green color represents low relative expression.
Validation of Altered miRNA Expressions by Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction Four miRNAs including mmu-miR-350-3p, mmumiR-202-3p, mmu-miR-711 and mmu-miR-30c-1-3p were randomly selected to further validate by qRT-PCR. The relative expression level of mmu-miR-350-3p and mmu-miR-202-3p were significantly increased to 1.518-fold and 3.731-fold in mice with OIR compared to control mice (P=0.0097 and P=0.0003, respectively; Figure 2), while that of mmu-miR-711 and mmu-miR-30c-1-3p was significantly decreased to 0.537-fold and 0.738-fold in mice with OIR compared to control mice (P=0.0006 and P=0.0121, respectively; Figure 2).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis of miRNA-Target Genes To gain insight into the possible roles of the altered miRNAs in OIR, we next predicted their potential target genes that may be involved in retinal neovascularization by 6 chosen miRNAs (mmu-miR-350-3p, mmu-miR-202-3p, mmumiR-711, mmu-miR-30c-1-3p, mmu-miR-201-3p and mmumiR-383-5p). Totally 1950 and 1030 target genes were identified, respectively, by using Targetscan7.1 and MirdbV5 database. By overlapping two sets of identified genes (Figure 3A), a total of 293 genes that shared in both databases were further identified. To illustrate the prediction of miRNA-gene interaction in a visualized form, miRNA-target gene network in mice with OIR was constructed (Figure 3B). The crossinteracting network showed that most of altered miRNAs can connect with other miRNAs based on their co-regulating genes, except for mmu-miR-711.
Table 1 miRNAs with significantly altered expression in retinas of mice with OIR identified by microarray
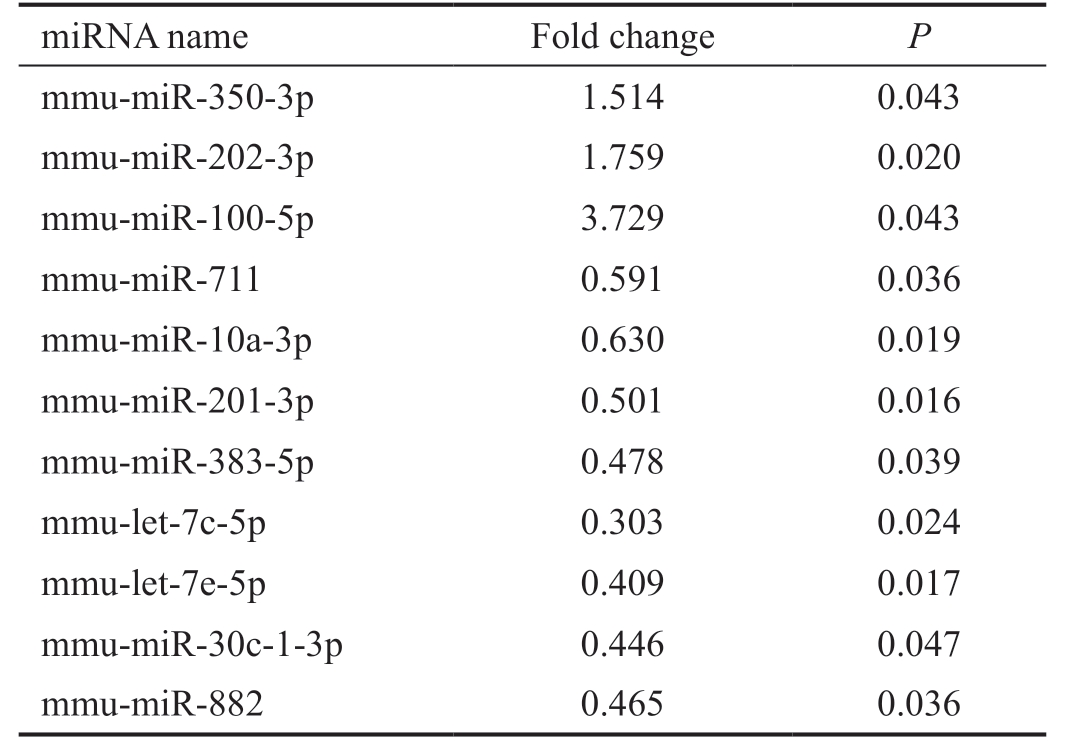
Data were filtered by fold change 1.5-fold up and down, and P<0.05.
miRNA name Fold change P mmu-miR-350-3p 1.514 0.043 mmu-miR-202-3p 1.759 0.020 mmu-miR-100-5p 3.729 0.043 mmu-miR-711 0.591 0.036 mmu-miR-10a-3p 0.630 0.019 mmu-miR-201-3p 0.501 0.016 mmu-miR-383-5p 0.478 0.039 mmu-let-7c-5p 0.303 0.024 mmu-let-7e-5p 0.409 0.017 mmu-miR-30c-1-3p 0.446 0.047 mmu-miR-882 0.465 0.036
The predicted target genes were subjected to GO classification and KEGG pathway enrichment analysis. Figure 4A demonstrates the top 10 enriched GO classifications in terms of biological process, cellular component and molecular function for identified target genes of altered miRNAs. The top 3 GO terms associated with biological process such as “cellular macromolecule metabolic process”, “regulation of biosynthetic process” and “nitrogen compound metabolic process”. The top 3 GO terms associated with cellular component include“intracellular”, “organelle” and “cell” part. The top 3 GO terms in molecular function are “binding”, “protein binding”and “catalytic activity”. As shown in Figure 4B, the KEGG pathways the predicted target genes indicate that the target genes regulated by those altered miRNAs mediate crosstalk between numerous pathways including Wnt signaling pathway,transcriptional misregulation in cancer, Mucin type O-glycan biosynthesis, and mitogen-activated protein kinase (MAPK)signaling pathway.
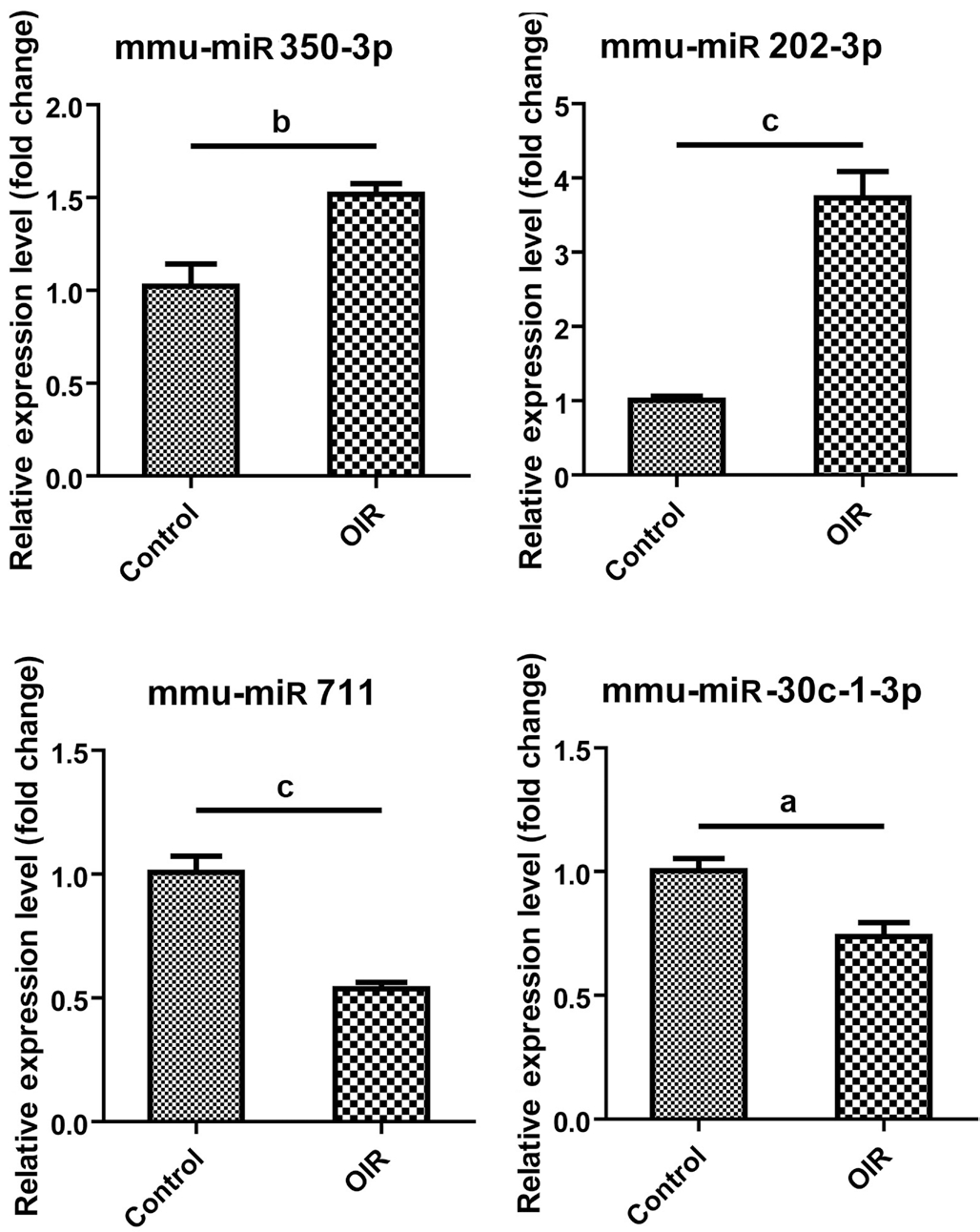
Figure 2 Validation of differential miRNA expression by qRTPCR Relative miRNA expression of mmu-miR-350-3p, mmu-miR-202-3p, mmu-miR-711, and mmu-miR-30c-1-3p in the retina from OIR and control mice. As compared to control, n=4/each group.aP<0.05, bP<0.01, cP<0.001.

Figure 3 The miRNA-target gene summary and the network analysis A: The Venn plot of the miRNA-target gene summary showed each predicted gene number in Targetscan7.1 and MirdbV5 databases and the overlapping gene number among these two prediction tools; B: The miRNA-target gene cross-interacting network. The network showed the direct interaction among the miRNAs and the target genes that may response to hypoxia and angiogenesis in OIR.
DISCUSSION
The present study examined retinal miRNA expression profile in mice with OIR through microarray and identified several upregulated and downregulated miRNAs that might contribute to pathological process of retinal neovascularization. Similar studies also profiled retinal miRNA expression and identified a number of altered miRNAs in mice with OIR[27-29]. Interestingly,the present study showed different results with identification of several altered miRNAs that were not been reported,including mmu-miR-350-3p, mmu-miR-202-3p, mmumiR-711, and mmu-miR-30c-1-3p, etc. Besides, other studies demonstrated the roles played by miRNAs such as miR-218[32],miR-155[33], miR-184[34] and miR-17 family[35]. Differences in the altered miRNAs identified among studies may relate to methodological differences between the arrays such as different miRNA primer alignments.
Macrophages are angiogenic-effecters in retinal neovascularization[36], and miR-350 plays an important role in macrophage apoptosis via negative regulation of PIK3R3 gene[37]. Our study showed that expression of miR-350-3p is significantly increased in mice with OIR, suggesting that miR-350-3p is likely to regulate macrophages apoptosis in retinal angiogenesis.Fibrosis is considered to be the later stage in retinal neovascularization[38] and miRNAs are involved in fibrosis[39-40].A study have demonstrated that fibrosis is suppressed in scleroderma by miR-202-3p via inhibition of MMP1, a profibrotic gene[41]. We showed that miR-202-3p is significantly upregulated in mice with OIR, indicating that miR-202-3p might affect fibrosis in retinal neovascularization through regulating its target genes.We also showed that several miRNAs, such as miR-711 and miR-30c-1-3p, were significantly downregulated in mice with OIR compared to room air controls. miR-711 was reported to inhibit angiopoietin-1, a well-known endothelial growth factor, through Akt pathway, and resulted in neuronal cell death[42]. Thus, miR-711 might be an inhibitor of retinal neovascularization through regulation of Akt pathway. miR-30c-1-3p is a functional miRNA in oxidized low-density lipoprotein-stimulated macrophages[43]. It might target interleukin-1β, a pro-inflammatory cytokine in acute gouty arthritis[44]. miR-30c-1-3p might contribute to the macrophage polarization and cytokine production so that it might also take part in the pathogenesis of angiogenesis.It has been reported that miRNAs play regulatory roles in regulation of VEGF expression. For instance, miRNA-181a attenuated ocular neovascularization through interfering with the expression of VEGF[45], and miRNA-16 regulated VEGF expression in retinal pigment epithelial cells[46].Although VEGF has been proved to be essential in retinal neovascularization, other molecules and pathways might also be involved in the pathogenesis. In the present study, GO and KEGG enrichment analyses suggested that these target gene enriched in cellular macromolecule metabolic process,regulation of biosynthetic process, and nitrogen compound metabolic process, which showed that the basic approach of synthesis and metabolism were changed. The reason might be that hypoxia causes physiological dysfunction, and leads to pathological neovascularization. Some important target genes are involved in our indicated pathways. For example, our bioinformatic analysis revealed that mmu-miR-350-3p target gene DKK2, and mmu-miR-30c-1-3p target gene GSK3B is key effectors in Wnt signaling pathway. Moreover, mmu-miR-30c-1-3p target genes IL1R1 and MAP3K3 are crucial factors in MAPK signaling pathway.
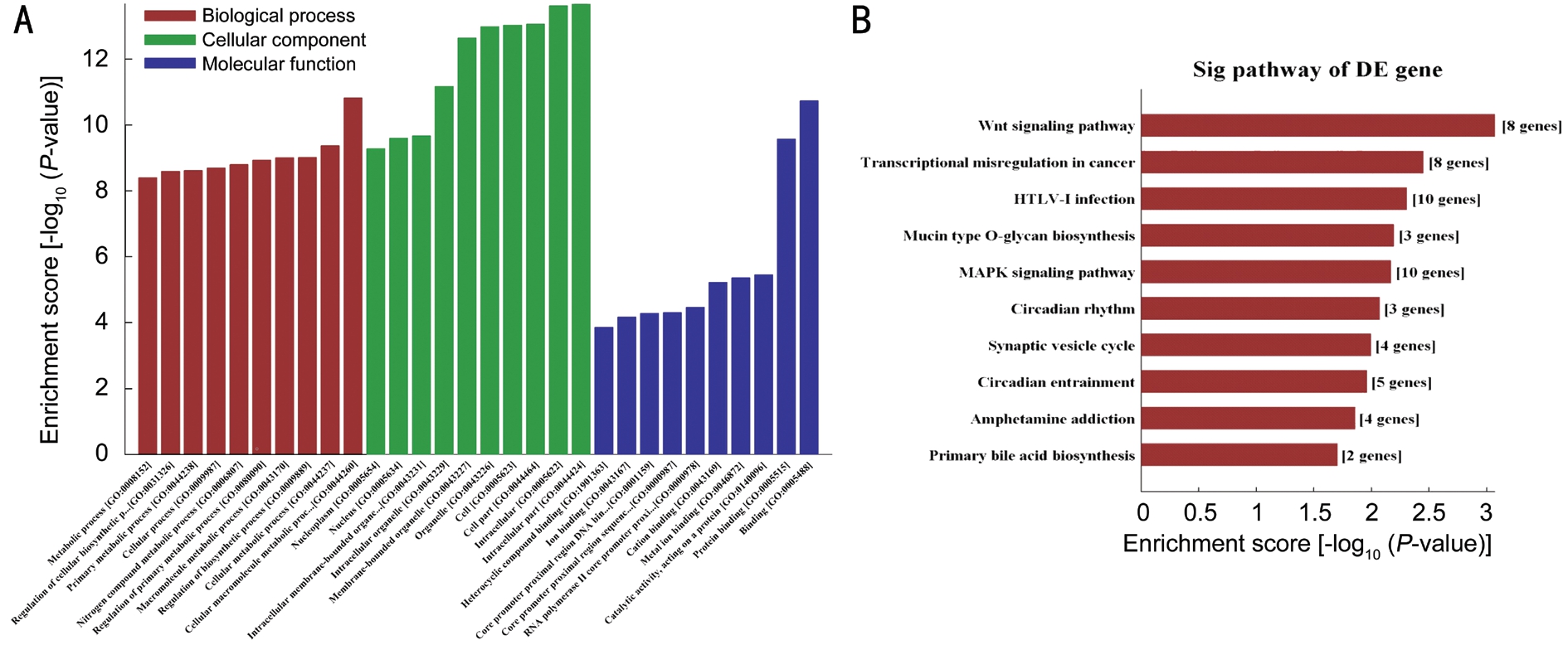
Figure 4 GO and KEGG pathway analyses of target genes of miRNAs The differential expressed miRNAs-target genes in OIR were subjected to GO (A) and KEGG pathway enrichment (B) analyses. A: Top 10 GO terms belongs to biological process, cellular component, and molecular function; B: Top 10 KEGG pathways among the miRNAs-target genes.
Our study also showed some limitations. Firstly, the data of microarray may not show all of the functional miRNAs. As a result, further effective detection methods such as Nextgeneration Sequence are needed for those miRNAs which expressed lower than the detective limitation. Moreover, not all identified miRNA have been validated by qRT-PCR, and the possible mechanism and effects of important target genes regulated by the altered miRNA have not been clarified in our study. Thus in vivo and in vitro studies should be done to explicit potential mechanisms in the mice with OIR.In conclusion, the study identified a number of altered miRNAs in mice with OIR and predicted the potential pathways and cellular function of those target genes of the miRNAs that involved in retinal neovascularization. Thus, identification of novel miRNAs or its target genes allows the revelation of the therapeutic targets and the potential approaches to therapies.
ACKNOWLEDGEMENTS
Foundations: Supported by National Natural Science Foundation of China (No.81700837; No.81800855); Natural Science Foundation of Hunan Province (No.2017JJ3452;No.2018JJ3765); Department of Science and Technology,Hunan (No.2015TP2007).
Conflicts of Interest: Zhang LS, None; Zhou YD, None;Peng YQ, None; Zeng HL, None; Yoshida S, None; Zhao TT, None.
1 Yoshida A, Yoshida S, Ishibashi T, Inomata H. Intraocular neovascularization. Histol Histopathol 1999;14(4):1287-1294.
2 Campochiaro PA. Ocular neovascularization. J Mol Med 2013;91(3):311-321.
3 Gariano RF, Gardner TW. Retinal angiogenesis in development and disease. Nature 2005;438(7070):960-966.
4 Osaadon P, Fagan XJ, Lifshitz T, Levy J. A review of anti-VEGF agents for proliferative diabetic retinopathy. Eye (Lond) 2014;28(5):510-520.
5 Rizzo S, Genovesi-Ebert F, Di Bartolo E, Vento A, Miniaci S, Williams G. Injection of intravitreal bevacizumab (Avastin) as a preoperative adjunct before vitrectomy surgery in the treatment of severe proliferative diabetic retinopathy (PDR). Graefes Arch Clin Exp Ophthalmol 2008;246(6):837-842.
6 Hosseini H, Khalili MR, Nowroozizadeh S. Intravitreal injection of bevacizumab (Avastin) for treatment of stage 3 retinopathy of prematurity in zone I or posterior zone II. Retina 2009;29(4):562.
7 Xu JJ, Li YM, Hong JX. Progress of anti-vascular endothelial growth factor therapy for ocular neovascular disease: benefits and challenges.Chin Med J 2014;127(8):1550-1557.
8 Kobayashi Y, Yoshida S, Zhou YD, et al. Tenascin-C promotes angiogenesis in fibrovascular membranes in eyes with proliferative diabetic retinopathy. Mol Vis 2016;22:436-445.
9 Nakama T, Yoshida S, Ishikawa K, et al. Different roles played by periostin splice variants in retinal neovascularization. Exp Eye Res 2016;153:133-140.
10 Connor KM, Krah NM, Dennison RJ, Aderman CM, Chen J, Guerin KI, Sapieha P, Stahl A, Willett KL, Smith LE. Quantification of oxygeninduced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat Protoc 2009;4(11):1565-1573.
11 Sato T, Kusaka S, Hashida N, Saishin Y, Fujikado T, Tano Y.Comprehensive gene-expression profile in murine oxygen-induced retinopathy. Br J Ophthalmol 2009;93(1):96-103.
12 Ishikawa K, Yoshida S, Kadota K, Nakamura T, Niiro H, Arakawa S,Yoshida A, Akashi K, Ishibashi T. Gene expression profile of hyperoxic and hypoxic retinas in a mouse model of oxygen-induced retinopathy.Invest Ophthalmol Vis Sci 2010;51(8):4307-4319.
13 Recchia FM, Xu LL, Penn JS, Boone B, Dexheimer PJ. Identification of genes and pathways involved in retinal neovascularization by microarray analysis of two animal models of retinal angiogenesis. Invest Ophthalmol Vis Sci 2010;51(2):1098-1105.
14 Zhou YD, Yoshida S, Nakao S, et al. M2 macrophages enhance pathological neovascularization in the mouse model of oxygen-induced retinopathy. Invest Ophthalmol Vis Sci 2015;56(8):4767-4777.
15 Nakama T, Yoshida S, Ishikawa K, et al. Therapeutic effect of novel single-stranded RNAi agent targeting periostin in eyes with retinal neovascularization. Mol Ther Nucleic Acids 2017;6:279-289.
16 Zhou YD, Yoshida S, Kubo Y, et al. Interleukin-12 inhibits pathological neovascularization in mouse model of oxygen-induced retinopathy. Sci Rep 2016;6:28140.
17 Huntzinger E, Izaurralde E. Gene silencing by microRNAs:contributions of translational repression and mRNA decay. Nat Rev Genet 2011;12(2):99-110.
18 Guo HL, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466(7308):835-840.
19 Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 2010;11(4):252-263.
20 Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers,functions and therapy. Trends Mol Med 2014;20(8):460-469.
21 Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011;469(7330):336-342.
22 Londin E, Loher P, Telonis AG, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA 2015;112(10):E1106-E1115.
23 He SK, Li XH, Chan N, Hinton DR. Review: Epigenetic mechanisms in ocular disease. Mol Vis 2013;19:665-674.
24 Zhou QY, Xiao X, Wang CK, Zhang XD, Li FZ, Zhou Y, Kijlstra A,Yang PZ. Decreased microRNA-155 expression in ocular behcet’s disease but not in vogt koyanagi harada syndrome. Invest Ophthalmol Vis Sci 2012;53(9):5665.
25 Hother C, Rasmussen PK, Joshi T, Reker D, Ralfkiær U, Workman CT, Heegaard S, Ralfkiær E, Grønbæk K. MicroRNA profiling in ocular adnexal lymphoma: a role for MYC and NFKB1 mediated dysregulation of microRNA expression in aggressive disease. Invest Ophthalmol Vis Sci 2013;54(8):5169-5175.
26 McArthur K, Feng B, Wu YX, Chen SL, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 2011;60(4):1314-1323.
27 Wang YP, Wu SY, Yang Y, Peng F, Li QT, Tian P, Xiang EY, Liang HL, Wang BB, Zhou XY, Huang H, Zhou XG. Differentially expressed miRNAs in oxygen-induced retinopathy newborn mouse models. Mol Med Rep 2017;15(1):146-152.
28 Liu CH, Wang ZX, Sun Y, SanGiovanni JP, Chen J. Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy.Sci Rep 2016;6:33947.
29 Shen JK, Yang XR, Xie B, Chen YJ, Swaim M, Hackett SF,Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther 2008;16(7):1208-1216.
30 Yamaji Y, Yoshida S, Ishikawa K, et al. TEM7 (PLXDC1) in neovascular endothelial cells of fibrovascular membranes from patients with proliferative diabetic retinopathy. Invest Ophthalmol Vis Sci 2008;49(7):3151-3157.
31 Ishikawa K, Yoshida S, Nakao S, Sassa Y, Asato R, Kohno R, Arima M, Kita T, Yoshida A, Ohuchida K, Ishibashi T. Bone marrow-derived monocyte lineage cells recruited by MIP-1β promote physiological revascularization in mouse model of oxygen-induced retinopathy. Lab Invest 2012;92(1):91-101.
32 Han S, Kong YC, Sun B, Han QH, Chen Y, Wang YC. microRNA-218 inhibits oxygen-induced retinal neovascularization via reducing the expression of roundabout 1. Chin Med J 2016;129(6):709-715.
33 Zhuang Z, Qin X, Hu H, Tian SY, Lu ZJ, Zhang TZ, Bai YL. Downregulation of microRNA-155 attenuates retinal neovascularization via the PI3K/Akt pathway. Mol Vis 2015;21:1173-1184.
34 Takahashi Y, Chen Q, Rajala RVS, Ma JX. MicroRNA-184 modulates canonical Wnt signaling through the regulation of frizzled-7 expression in the retina with ischemia-induced neovascularization. FEBS Lett 2015;589(10):1143-1149.
35 Nunes DN, Dias-Neto E, Cardó-Vila M, et al. Synchronous downmodulation of miR-17 family members is an early causative event in the retinal angiogenic switch. Proc Natl Acad Sci USA 2015;112(12):3770-3775.
36 Zhou YD, Yoshida S, Peng YQ, Kobayashi Y, Zhang LS, Tang LS.Diverse roles of macrophages in intraocular neovascular diseases: a review. Int J Ophthalmol 2017;10(12):1902-1908.
37 Sui J, Fu YY, Zhang YQ, Ma SM, Yin LH, Pu YP, Liang GY.Molecular mechanism for miR-350 in regulating of titanium dioxide nanoparticles in macrophage RAW264.7 cells. Chem Biol Interact 2018;280:77-85.
38 Friedlander M. Fibrosis and diseases of the eye. J Clin Invest 2007;117(3):576-586.
39 van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH,Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008;105(35):13027-13032.
40 Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J,Zern MA. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol 2010;298(1):G101-G106.
41 Zhou B, Zhu HL, Luo H, Gao SM, Dai XD, Li YS, Zuo XX.MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed Pharmacother 2017;87:412-418.
42 Sabirzhanov B, Faden AI, Aubrecht T, Henry R, Glaser E, Stoica BA.MicroRNA-711-induced downregulation of angiopoietin-1 mediates neuronal cell death. J Neurotrauma 2018;35(20):2462-2481.
43 Li XK, Feng SY, Luo Y, Long KR, Lin ZH, Ma JD, Jiang AN, Jin L,Tang QZ, Li MZ, Wang X. Expression profiles of microRNAs in oxidized low-density lipoprotein-stimulated RAW 264.7 cells. In Vitro Cell Dev Biol Anim 2018;54(2):99-110.
44 Zhou WD, Wang Y, Wu RF, He Y, Su Q, Shi GX. MicroRNA-488 and-920 regulate the production of proinflammatory cytokines in acute gouty arthritis. Arthritis Res Ther 2017;19(1):203.
45 Yang C, Tahiri H, Cai C, Gu MQ, Gagnon C, Hardy P. microRNA-181a inhibits ocular neovascularization by interfering with vascular endothelial growth factor expression. Cardiovasc Ther 2018;36(3):e12329.
46 Huang JF, Wang YQ, Wang LN, Pan Y, Chen T. MicroRNA-16 inhibits hypoxia-induced vascular endothelial growth factor expression in ARPE-19 cells. Cutan Ocul Toxicol 2018;37(3):228-232.